7D4R
 
 | | SpuA native structure | | Descriptor: | MAGNESIUM ION, Probable glutamine amidotransferase | | Authors: | Chen, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-09-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and mechanism of the gamma-glutamyl-gamma-aminobutyrate hydrolase SpuA from Pseudomonas aeruginosa.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7D50
 
 | | SpuA mutant - H221N with glutamyl-thioester | | Descriptor: | MAGNESIUM ION, Probable glutamine amidotransferase | | Authors: | Chen, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-09-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure and mechanism of the gamma-glutamyl-gamma-aminobutyrate hydrolase SpuA from Pseudomonas aeruginosa.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7D53
 
 | | SpuA mutant - H221N with Glu | | Descriptor: | GLUTAMIC ACID, MAGNESIUM ION, Probable glutamine amidotransferase | | Authors: | Chen, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-09-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and mechanism of the gamma-glutamyl-gamma-aminobutyrate hydrolase SpuA from Pseudomonas aeruginosa.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7D9F
 
 | | SpdH Spermidine dehydrogenase SeMet Structure | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Che, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-10-13 | | Release date: | 2021-11-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of Pseudomonas aeruginosa spermidine dehydrogenase: a polyamine oxidase with a novel heme-binding fold.
Febs J., 289, 2022
|
|
7E7G
 
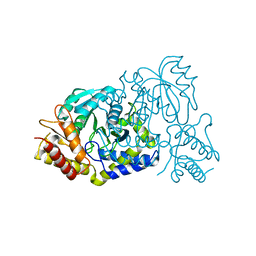 | |
5M1S
 
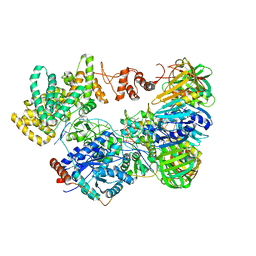 | | Cryo-EM structure of the E. coli replicative DNA polymerase-clamp-exonuclase-theta complex bound to DNA in the editing mode | | Descriptor: | DNA Primer Strand, DNA Template Strand, DNA polymerase III subunit alpha, ... | | Authors: | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | Deposit date: | 2016-10-10 | | Release date: | 2017-01-18 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Self-correcting mismatches during high-fidelity DNA replication.
Nat. Struct. Mol. Biol., 24, 2017
|
|
7EPO
 
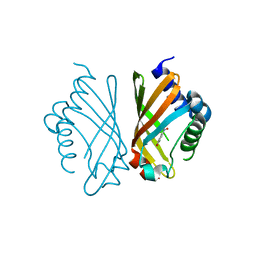 | | Ketosteroid Isomerase KSI with 5-nitrobenzoxazole (5NBI) | | Descriptor: | 5-nitro-1,2-benzoxazole, SnoaL-like domain-containing protein | | Authors: | Liang, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2021-04-27 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural characterization and Kemp eliminase activity of the Mycobacterium smegmatis Ketosteroid Isomerase.
Biochem.Biophys.Res.Commun., 560, 2021
|
|
7EPN
 
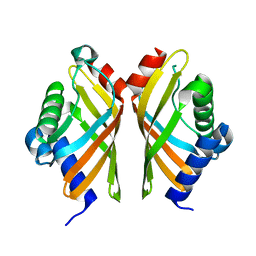 | | Ketosteroid Isomerase KSI native | | Descriptor: | SnoaL-like domain-containing protein | | Authors: | Liang, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2021-04-27 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization and Kemp eliminase activity of the Mycobacterium smegmatis Ketosteroid Isomerase.
Biochem.Biophys.Res.Commun., 560, 2021
|
|
7D54
 
 | | Crstal structure MsGATase with Gln | | Descriptor: | GLUTAMINE, Glutamine amidotransferase class-I | | Authors: | Chen, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-09-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and mechanism of the gamma-glutamyl-gamma-aminobutyrate hydrolase SpuA from Pseudomonas aeruginosa.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7D9I
 
 | | SpdH Spermidine dehydrogenase D282A mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROTOPORPHYRIN IX CONTAINING FE, Spermidine dehydrogenase, ... | | Authors: | Che, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-10-13 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Pseudomonas aeruginosa spermidine dehydrogenase: a polyamine oxidase with a novel heme-binding fold.
Febs J., 289, 2022
|
|
7D9H
 
 | | SpdH Spermidine dehydrogenase N33 truncation structure | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Che, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-10-13 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure of Pseudomonas aeruginosa spermidine dehydrogenase: a polyamine oxidase with a novel heme-binding fold.
Febs J., 289, 2022
|
|
7D9J
 
 | | SpdH Spermidine dehydrogenase Y443A mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Che, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-10-13 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure of Pseudomonas aeruginosa spermidine dehydrogenase: a polyamine oxidase with a novel heme-binding fold.
Febs J., 289, 2022
|
|
7D9G
 
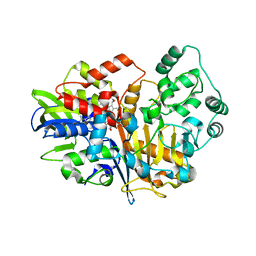 | | SpdH Spermidine dehydrogenase native structure | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROTOPORPHYRIN IX CONTAINING FE, Spermidine dehydrogenase, ... | | Authors: | Che, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-10-13 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Pseudomonas aeruginosa spermidine dehydrogenase: a polyamine oxidase with a novel heme-binding fold.
Febs J., 289, 2022
|
|
5A2S
 
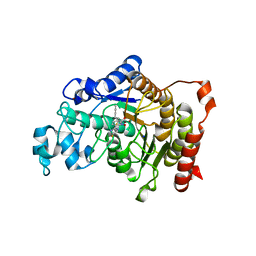 | | Potent, selective and CNS-penetrant tetrasubstituted cyclopropane class IIa histone deacetylase (HDAC) inhibitors | | Descriptor: | (1S,2S,3S)-1-fluoranyl-2-[4-(5-fluoranylpyrimidin-2-yl)phenyl]-N-oxidanyl-3-phenyl-cyclopropane-1-carboxamide, HISTONE DEACETYLASE 4, SODIUM ION, ... | | Authors: | Luckhurst, C.A, Breccia, P, Stott, A.J, Aziz, O, Birch, H, Burli, R.W, Hughes, S, Jarvis, R.E, Lamers, M, Leonard, P, Matthews, K.L, McAllister, G, Pollack, S, Saville-Stones, E, Wishart, G, Yates, D, Dominguez, C. | | Deposit date: | 2015-05-22 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Potent, Selective, and Cns-Penetrant Tetrasubstituted Cyclopropane Class Iia Histone Deacetylase (Hdac) Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
5FDL
 
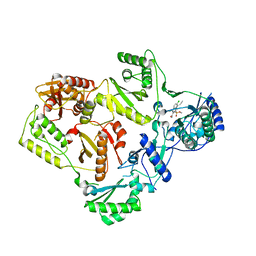 | | Crystal Structure of K103N/Y181C Mutant HIV-1 Reverse Transcriptase (RT) in Complex with IDX899 | | Descriptor: | P51 Reverse transcriptase, P66 Reverse transcriptase, methyl (R)-(2-carbamoyl-5-chloro-1H-indol-3-yl)[3-(2-cyanoethyl)-5-methylphenyl]phosphinate | | Authors: | Dousson, C.B, Alexandre, F.-R, Convard, T, Fisher, M, Lamers, M.B.A.C, Leonard, P.M. | | Deposit date: | 2015-12-16 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery of the Aryl-phospho-indole IDX899, a Highly Potent Anti-HIV Non-nucleoside Reverse Transcriptase Inhibitor.
J.Med.Chem., 59, 2016
|
|
5CI9
 
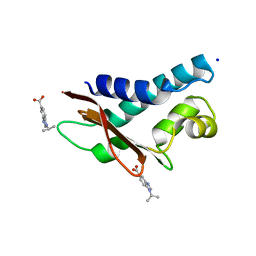 | | Crystal structure of human Tob in complex with inhibitor fragment 6 | | Descriptor: | 1-(propan-2-yl)-1H-benzimidazole-5-carboxylic acid, Protein Tob1, SODIUM ION | | Authors: | Bai, Y, Tashiro, S, Nagatoishi, S, Suzuki, T, Tsumoto, K, Bartlam, M, Yamamoto, T. | | Deposit date: | 2015-07-11 | | Release date: | 2015-11-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for inhibition of the Tob-CNOT7 interaction by a fragment screening approach
Protein Cell, 6, 2015
|
|
5CI8
 
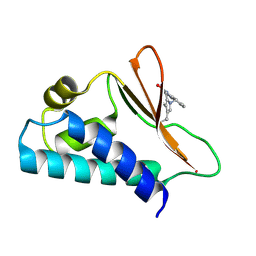 | | Crystal structure of human Tob in complex with inhibitor fragment 1 | | Descriptor: | Protein Tob1, pyrrolo[1,2-a]quinoxalin-4(5H)-one | | Authors: | Bai, Y, Tashiro, S, Nagatoishi, S, Suzuki, T, Tsumoto, K, Bartlam, M, Yamamoto, T. | | Deposit date: | 2015-07-11 | | Release date: | 2015-11-18 | | Last modified: | 2015-12-09 | | Method: | X-RAY DIFFRACTION (2.328 Å) | | Cite: | Structural basis for inhibition of the Tob-CNOT7 interaction by a fragment screening approach
Protein Cell, 6, 2015
|
|
5YKR
 
 | |
5YKT
 
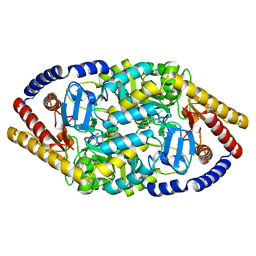 | |
4ZHU
 
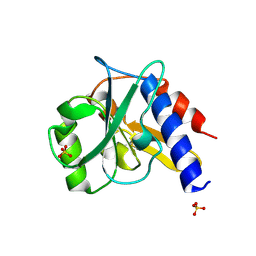 | | Crystal structure of a bacterial repressor protein | | Descriptor: | SULFATE ION, YfiR | | Authors: | Li, S, Li, T, Wang, Y, Bartlam, M. | | Deposit date: | 2015-04-27 | | Release date: | 2016-04-27 | | Method: | X-RAY DIFFRACTION (2.3968 Å) | | Cite: | Structural insights into YfiR sequestering by YfiB in Pseudomonas aeruginosa PAO1
Sci Rep, 5, 2015
|
|
4ZHW
 
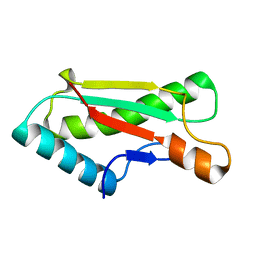 | |
5DV2
 
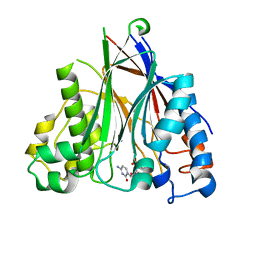 | |
4ZHY
 
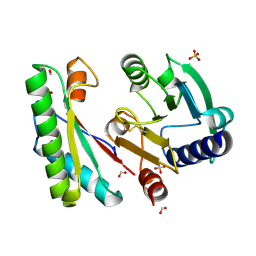 | | Crystal structure of a bacterial signalling complex | | Descriptor: | FORMIC ACID, SULFATE ION, YfiB, ... | | Authors: | Li, S, Li, T, Wang, Y, Bartlam, M. | | Deposit date: | 2015-04-27 | | Release date: | 2016-04-27 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | Structural insights into YfiR sequestering by YfiB in Pseudomonas aeruginosa PAO1
Sci Rep, 5, 2015
|
|
4ZHV
 
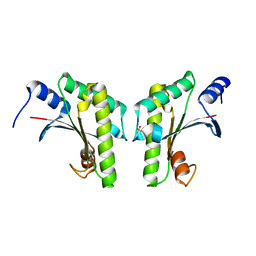 | | Crystal structure of a bacterial signalling protein | | Descriptor: | SULFATE ION, YfiB | | Authors: | Li, S, Li, T, Wang, Y, Bartlam, M. | | Deposit date: | 2015-04-27 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.585 Å) | | Cite: | Structural insights into YfiR sequestering by YfiB in Pseudomonas aeruginosa PAO1
Sci Rep, 5, 2015
|
|
7PU7
 
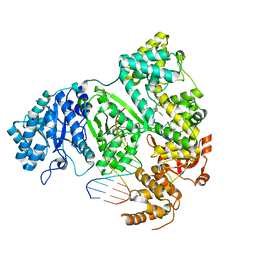 | | DNA polymerase from M. tuberculosis | | Descriptor: | DNA polymerase III subunit alpha, Template, ZINC ION, ... | | Authors: | Borsellini, A, Lamers, M.H. | | Deposit date: | 2021-09-28 | | Release date: | 2022-02-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | DNA-Dependent Binding of Nargenicin to DnaE1 Inhibits Replication in Mycobacterium tuberculosis.
Acs Infect Dis., 8, 2022
|
|
