7UHL
 
 | | Time-Resolved Structure of Metallo Beta-Lactamase L1 in a Complex with Non-Hydrolyzed Moxalactam (100 ms Snapshot) | | Descriptor: | (1R,6R,7R)-7-[(2R)-2-carboxypropanamido]-7-methoxy-3-methyl-8-oxo-5-oxa-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylic acid, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Henning, R, Maltseva, N, Endres, M, Babnigg, G, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-03-27 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Time-resolved beta-lactam cleavage by L1 metallo-beta-lactamase.
Nat Commun, 13, 2022
|
|
7UHQ
 
 | | Time-Resolved Structure of Metallo Beta-Lactamase L1 in a Complex with Cleaved Moxalactam (4000 ms Snapshot) | | Descriptor: | (2R)-2-[(R)-carboxy{[(2R)-2-carboxy-2-(4-hydroxyphenyl)acetyl]amino}methoxymethyl]-5-{[(1-methyl-1H-tetrazol-5-yl)sulfanyl]methyl}-3,6-dihydro-2H-1,3-oxazine-4-carboxylic acid, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Henning, R, Maltseva, N, Endres, M, Babnigg, G, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-03-27 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Time-resolved beta-lactam cleavage by L1 metallo-beta-lactamase.
Nat Commun, 13, 2022
|
|
7UHR
 
 | | Time-Resolved Structure of Metallo Beta-Lactamase L1 Before Reaction (Dark-Set) | | Descriptor: | Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Henning, R, Maltseva, N, Endres, M, Babnigg, G, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-03-27 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Time-resolved beta-lactam cleavage by L1 metallo-beta-lactamase.
Nat Commun, 13, 2022
|
|
7UHS
 
 | | SSX Structure of Metallo Beta-Lactamase L1 with Two Water Molecules in the Active Site | | Descriptor: | Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase) | | Authors: | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Henning, R, Maltseva, N, Endres, M, Babnigg, G, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-03-27 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Time-resolved beta-lactam cleavage by L1 metallo-beta-lactamase.
Nat Commun, 13, 2022
|
|
3EG5
 
 | | Crystal structure of MDIA1-TSH GBD-FH3 in complex with CDC42-GMPPNP | | Descriptor: | Cell division control protein 42 homolog, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Lammers, M, Meyer, S, Kuehlmann, D, Wittinghofer, A. | | Deposit date: | 2008-09-10 | | Release date: | 2008-10-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Specificity of Interactions between mDia Isoforms and Rho Proteins
J.Biol.Chem., 283, 2008
|
|
1VE6
 
 | | Crystal structure of an acylpeptide hydrolase/esterase from Aeropyrum pernix K1 | | Descriptor: | Acylamino-acid-releasing enzyme, GLYCEROL, octyl beta-D-glucopyranoside | | Authors: | Bartlam, M, Wang, G, Gao, R, Yang, H, Zhao, X, Xie, G, Cao, S, Feng, Y, Rao, Z. | | Deposit date: | 2004-03-27 | | Release date: | 2004-11-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an acylpeptide hydrolase/esterase from Aeropyrum pernix K1
STRUCTURE, 12, 2004
|
|
1VE7
 
 | | Crystal structure of an acylpeptide hydrolase/esterase from Aeropyrum pernix K1 in complex with p-nitrophenyl phosphate | | Descriptor: | 4-NITROPHENYL PHOSPHATE, Acylamino-acid-releasing enzyme, GLYCEROL | | Authors: | Bartlam, M, Wang, G, Gao, R, Yang, H, Zhao, X, Xie, G, Cao, S, Feng, Y, Rao, Z. | | Deposit date: | 2004-03-27 | | Release date: | 2004-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of an acylpeptide hydrolase/esterase from Aeropyrum pernix K1
STRUCTURE, 12, 2004
|
|
2BAP
 
 | |
1W7A
 
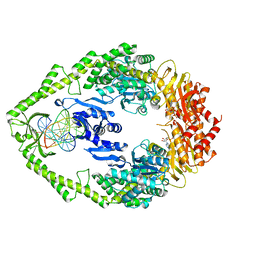 | | ATP bound MutS | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP *CP*AP*CP*CP*AP*GP*TP*GP*TP*CP*AP*GP*CP*GP*TP*CP*CP*TP* AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP *GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*TP*TP*GP*GP*CP*AP*GP* CP*T)-3', ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Lamers, M.H, Georgijevic, D, Lebbink, J, Winterwerp, H.H.K, Agianian, B, de Wind, N, Sixma, T.K. | | Deposit date: | 2004-08-31 | | Release date: | 2004-09-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | ATP Increases the Affinity between Muts ATPase Domains: Implications for ATP Hydrolysis and Conformational Changes
J.Biol.Chem., 279, 2004
|
|
1E3M
 
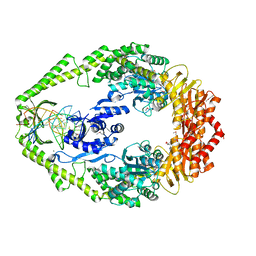 | | The crystal structure of E. coli MutS binding to DNA with a G:T mismatch | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP*CP*AP*CP*CP*AP* GP*TP*GP*TP*CP*AP*GP*CP*GP*TP*CP*CP*TP*AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP*GP*AP*CP*AP*CP* TP*GP*GP*TP*GP*CP*TP*TP*GP*GP*CP*AP*GP*CP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Lamers, M.H, Perrakis, A, Enzlin, J.H, Winterwerp, H.H.K, De Wind, N, Sixma, T.K. | | Deposit date: | 2000-06-19 | | Release date: | 2000-11-01 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of DNA Mismatch Repair Protein Muts Binding to a G X T Mismatch
Nature, 407, 2000
|
|
8B0I
 
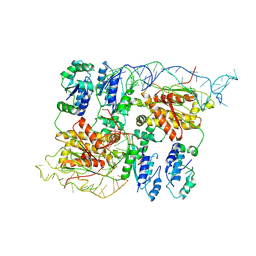 | | CryoEM structure of bacterial RapZ.GlmZ complex central to the control of cell envelope biogenesis | | Descriptor: | GlmZ small regulatory RNA, RNase adapter protein RapZ | | Authors: | Islam, M.S, Hardwick, H.W, Chirgadze, D.Y, Luisi, B.F. | | Deposit date: | 2022-09-07 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.28 Å) | | Cite: | Structure of a bacterial ribonucleoprotein complex central to the control of cell envelope biogenesis.
Embo J., 42, 2023
|
|
8B0J
 
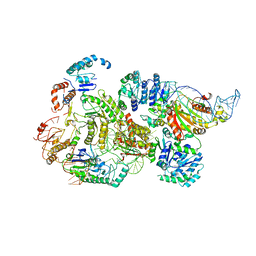 | | CryoEM structure of bacterial RNaseE.RapZ.GlmZ complex central to the control of cell envelope biogenesis | | Descriptor: | GlmZ small RNA, RNase adapter protein RapZ, Ribonuclease E | | Authors: | Islam, M.S, Hardwick, H.W, Chirgadze, D.Y, Luisi, B.F. | | Deposit date: | 2022-09-07 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Structure of a bacterial ribonucleoprotein complex central to the control of cell envelope biogenesis.
Embo J., 42, 2023
|
|
7OTO
 
 | | The structure of MutS bound to two molecules of AMPPNP | | Descriptor: | DNA mismatch repair protein MutS, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Lamers, M.H, Borsellini, A, Friedhoff, P, Kunetsky, V. | | Deposit date: | 2021-06-10 | | Release date: | 2022-01-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryogenic electron microscopy structures reveal how ATP and DNA binding in MutS coordinates sequential steps of DNA mismatch repair.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7OU4
 
 | | The structure of MutS bound to one molecule of ATP and one molecule of ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA mismatch repair protein MutS, ... | | Authors: | Lamers, M.H, Borsellini, A, Friedhoff, P, Kunetsky, V. | | Deposit date: | 2021-06-11 | | Release date: | 2022-01-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryogenic electron microscopy structures reveal how ATP and DNA binding in MutS coordinates sequential steps of DNA mismatch repair.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7OU0
 
 | | The structure of MutS bound to two molecules of ADP-Vanadate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA mismatch repair protein MutS, MAGNESIUM ION, ... | | Authors: | Lamers, M.H, Borsellini, A, Friedhoff, P, Kunetsky, V. | | Deposit date: | 2021-06-10 | | Release date: | 2022-01-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryogenic electron microscopy structures reveal how ATP and DNA binding in MutS coordinates sequential steps of DNA mismatch repair.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7OU2
 
 | | The structure of MutS bound to two molecules of ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA mismatch repair protein MutS | | Authors: | Lamers, M.H, Borsellini, A, Friedhoff, P, Kunetsky, V. | | Deposit date: | 2021-06-11 | | Release date: | 2022-01-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cryogenic electron microscopy structures reveal how ATP and DNA binding in MutS coordinates sequential steps of DNA mismatch repair.
Nat.Struct.Mol.Biol., 29, 2022
|
|
3GI6
 
 | | Crystal structure of protease inhibitor, AD78 in complex with wild type HIV-1 protease | | Descriptor: | (5S)-N-[(1S,2R)-2-Hydroxy-3-[[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino]-1-(phenylmethyl)propyl]-2-oxo-3-[3-(trif luoromethyl)phenyl]-5-oxazolidinecarboxamide, PHOSPHATE ION, Protease | | Authors: | Nalam, M.N.L, Schiffer, C.A. | | Deposit date: | 2009-03-05 | | Release date: | 2010-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Evaluating the substrate-envelope hypothesis: structural analysis of novel HIV-1 protease inhibitors designed to be robust against drug resistance.
J.Virol., 84, 2010
|
|
3GI5
 
 | | Crystal structure of protease inhibitor, KB62 in complex with wild type HIV-1 protease | | Descriptor: | (5S)-3-(3-Acetylphenyl)-N-[(1S,2R)-3-[(1,3-benzodioxol-5-ylsulfonyl)(2-methylpropyl)amino]-2-hydroxy-1-(phenylmethyl)pr opyl]-2-oxo-5-oxazolidinecarboxamide, PHOSPHATE ION, Protease | | Authors: | Nalam, M.N.L, Schiffer, C.A. | | Deposit date: | 2009-03-05 | | Release date: | 2010-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evaluating the substrate-envelope hypothesis: structural analysis of novel HIV-1 protease inhibitors designed to be robust against drug resistance.
J.Virol., 84, 2010
|
|
3MXD
 
 | | Crystal structure of HIV-1 protease inhibitor KC53 in complex with wild-type protease | | Descriptor: | (5S)-N-{(1S,2R)-3-[(1,3-benzodioxol-5-ylsulfonyl)(2-methylpropyl)amino]-1-benzyl-2-hydroxypropyl}-3-(2-hydroxyphenyl)-2 -oxo-1,3-oxazolidine-5-carboxamide, ACETATE ION, HIV-1 protease, ... | | Authors: | Nalam, M.N.L, Schiffer, C.A. | | Deposit date: | 2010-05-07 | | Release date: | 2010-11-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Based Design, Synthesis, and Structure-Activity Relationship Studies of HIV-1 Protease Inhibitors Incorporating Phenyloxazolidinones.
J.Med.Chem., 53, 2010
|
|
3MXE
 
 | | Crystal structure of HIV-1 protease inhibitor, KC32 complexed with wild-type protease | | Descriptor: | (5S)-N-{(1S,2R)-3-[(1,3-benzothiazol-6-ylsulfonyl)(2-methylpropyl)amino]-1-benzyl-2-hydroxypropyl}-2-oxo-3-[2-(trifluoromethyl)phenyl]-1,3-oxazolidine-5-carboxamide, ACETATE ION, HIV-1 protease, ... | | Authors: | Nalam, M.N.L, Schiffer, C.A. | | Deposit date: | 2010-05-07 | | Release date: | 2010-11-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Design, Synthesis, and Structure-Activity Relationship Studies of HIV-1 Protease Inhibitors Incorporating Phenyloxazolidinones.
J.Med.Chem., 53, 2010
|
|
3GI4
 
 | | Crystal structure of protease inhibitor, KB60 in complex with wild type HIV-1 protease | | Descriptor: | 5S)-N-[(1S,2R)-3-[(1,3-Benzodioxol-5-ylsulfonyl)(2-methylpropyl)amino]-2-hydroxy-1-(phenylmethyl)propyl]-2-oxo-3-[3-(tr ifluoromethyl)phenyl]-5-oxazolidinecarboxamide, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Nalam, M.N.L, Schiffer, C.A. | | Deposit date: | 2009-03-05 | | Release date: | 2010-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Evaluating the substrate-envelope hypothesis: structural analysis of novel HIV-1 protease inhibitors designed to be robust against drug resistance.
J.Virol., 84, 2010
|
|
2QI4
 
 | | Crystal structure of protease inhibitor, MIT-2-AD93 in complex with wild type HIV-1 protease | | Descriptor: | ACETATE ION, N-[(1S,2R)-3-{(1,3-BENZOTHIAZOL-6-YLSULFONYL)[(2S)-2-METHYLBUTYL]AMINO}-1-BENZYL-2-HYDROXYPROPYL]-3-HYDROXYBENZAMIDE, PHOSPHATE ION, ... | | Authors: | Nalam, M.N.L, Schiffer, C.A. | | Deposit date: | 2007-07-03 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | HIV-1 protease inhibitors from inverse design in the substrate envelope exhibit subnanomolar binding to drug-resistant variants.
J.Am.Chem.Soc., 130, 2008
|
|
2QI3
 
 | | Crystal structure of protease inhibitor, MIT-2-AD94 in complex with wild type HIV-1 protease | | Descriptor: | (2S)-N-[(1S,2R)-3-{(1,3-BENZOTHIAZOL-6-YLSULFONYL)[(2S)-2-METHYLBUTYL]AMINO}-1-BENZYL-2-HYDROXYPROPYL]-2-HYDROXY-3-METHYLBUTANAMIDE, PHOSPHATE ION, Protease | | Authors: | Nalam, M.N.L, Schiffer, C.A. | | Deposit date: | 2007-07-03 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | HIV-1 protease inhibitors from inverse design in the substrate envelope exhibit subnanomolar binding to drug-resistant variants.
J.Am.Chem.Soc., 130, 2008
|
|
8BAG
 
 | |
8BAE
 
 | |
