5JB7
 
 | |
5XX5
 
 | | A BPTI-[5,55] variant with C14GA38I mutations | | Descriptor: | Pancreatic trypsin inhibitor, SULFATE ION | | Authors: | Islam, M.M. | | Deposit date: | 2017-07-01 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Hydrophobic surface residues can stabilize a protein through improved water-protein interactions.
Febs J., 2019
|
|
5XX2
 
 | | A BPTI-[5,55] variant with C14GA38L mutations | | Descriptor: | Pancreatic trypsin inhibitor, SULFATE ION | | Authors: | Islam, M.M. | | Deposit date: | 2017-07-01 | | Release date: | 2018-07-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Hydrophobic surface residues can stabilize a protein through improved water-protein interactions.
Febs J., 2019
|
|
5XX4
 
 | | A BPTI-[5,55] variant with C14GA38K mutations | | Descriptor: | Pancreatic trypsin inhibitor, SULFATE ION | | Authors: | Islam, M.M. | | Deposit date: | 2017-07-01 | | Release date: | 2018-07-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Hydrophobic surface residues can stabilize a protein through improved water-protein interactions.
Febs J., 2019
|
|
5XX3
 
 | | A BPTI-[5,55] variant with C14GA38G mutations | | Descriptor: | Pancreatic trypsin inhibitor, SULFATE ION | | Authors: | Islam, M.M. | | Deposit date: | 2017-07-01 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Hydrophobic surface residues can stabilize a protein through improved water-protein interactions.
Febs J., 2019
|
|
5XX8
 
 | |
5XX7
 
 | |
5XX6
 
 | |
3L3T
 
 | | Human mesotrypsin complexed with amyloid precursor protein inhibitor variant (APPIR15K) | | Descriptor: | CALCIUM ION, FORMIC ACID, PRSS3 protein, ... | | Authors: | Salameh, M.A, Soares, A.S, Radisky, E.S. | | Deposit date: | 2009-12-17 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.378 Å) | | Cite: | Determinants of affinity and proteolytic stability in interactions of Kunitz family protease inhibitors with mesotrypsin.
J.Biol.Chem., 285, 2010
|
|
3L33
 
 | | Human mesotrypsin complexed with amyloid precursor protein inhibitor(APPI) | | Descriptor: | Amyloid beta A4 protein, CALCIUM ION, FORMIC ACID, ... | | Authors: | Salameh, M.A, Soares, A.S, Radisky, E.S. | | Deposit date: | 2009-12-16 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Determinants of affinity and proteolytic stability in interactions of Kunitz family protease inhibitors with mesotrypsin.
J.Biol.Chem., 285, 2010
|
|
1VE7
 
 | | Crystal structure of an acylpeptide hydrolase/esterase from Aeropyrum pernix K1 in complex with p-nitrophenyl phosphate | | Descriptor: | 4-NITROPHENYL PHOSPHATE, Acylamino-acid-releasing enzyme, GLYCEROL | | Authors: | Bartlam, M, Wang, G, Gao, R, Yang, H, Zhao, X, Xie, G, Cao, S, Feng, Y, Rao, Z. | | Deposit date: | 2004-03-27 | | Release date: | 2004-11-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of an acylpeptide hydrolase/esterase from Aeropyrum pernix K1
STRUCTURE, 12, 2004
|
|
8BAG
 
 | |
8BAF
 
 | |
8BAE
 
 | |
6WIQ
 
 | | Crystal structure of the co-factor complex of NSP7 and the C-terminal domain of NSP8 from SARS CoV-2 | | Descriptor: | Non-structural protein 7, Non-structural protein 8 | | Authors: | Wilamowski, M, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Transient and stabilized complexes of Nsp7, Nsp8, and Nsp12 in SARS-CoV-2 replication.
Biophys.J., 120, 2021
|
|
7OU4
 
 | | The structure of MutS bound to one molecule of ATP and one molecule of ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA mismatch repair protein MutS, ... | | Authors: | Lamers, M.H, Borsellini, A, Friedhoff, P, Kunetsky, V. | | Deposit date: | 2021-06-11 | | Release date: | 2022-01-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryogenic electron microscopy structures reveal how ATP and DNA binding in MutS coordinates sequential steps of DNA mismatch repair.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7OU2
 
 | | The structure of MutS bound to two molecules of ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA mismatch repair protein MutS | | Authors: | Lamers, M.H, Borsellini, A, Friedhoff, P, Kunetsky, V. | | Deposit date: | 2021-06-11 | | Release date: | 2022-01-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cryogenic electron microscopy structures reveal how ATP and DNA binding in MutS coordinates sequential steps of DNA mismatch repair.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7OU0
 
 | | The structure of MutS bound to two molecules of ADP-Vanadate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA mismatch repair protein MutS, MAGNESIUM ION, ... | | Authors: | Lamers, M.H, Borsellini, A, Friedhoff, P, Kunetsky, V. | | Deposit date: | 2021-06-10 | | Release date: | 2022-01-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryogenic electron microscopy structures reveal how ATP and DNA binding in MutS coordinates sequential steps of DNA mismatch repair.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7OTO
 
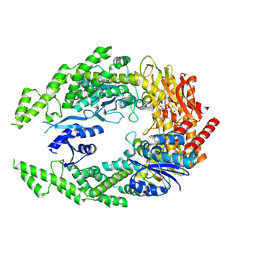 | | The structure of MutS bound to two molecules of AMPPNP | | Descriptor: | DNA mismatch repair protein MutS, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Lamers, M.H, Borsellini, A, Friedhoff, P, Kunetsky, V. | | Deposit date: | 2021-06-10 | | Release date: | 2022-01-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryogenic electron microscopy structures reveal how ATP and DNA binding in MutS coordinates sequential steps of DNA mismatch repair.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6WTC
 
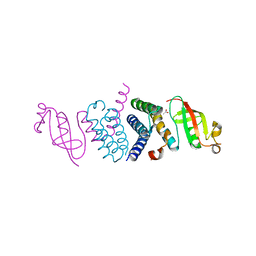 | | Crystal Structure of the Second Form of the Co-factor Complex of NSP7 and the C-terminal Domain of NSP8 from SARS CoV-2 | | Descriptor: | ACETIC ACID, Non-structural protein 7, Non-structural protein 8 | | Authors: | Wilamowski, M, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-02 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Second Form of the Co-factor Complex of NSP7 and the C-terminal Domain of NSP8 from SARS CoV-2
To Be Published
|
|
6XIP
 
 | | The 1.5 A Crystal Structure of the Co-factor Complex of NSP7 and the C-terminal Domain of NSP8 from SARS CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, Non-structural protein 7, Non-structural protein 8 | | Authors: | Wilamowski, M, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-20 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Transient and stabilized complexes of Nsp7, Nsp8, and Nsp12 in SARS-CoV-2 replication.
Biophys.J., 120, 2021
|
|
6XKM
 
 | | Room Temperature Structure of SARS-CoV-2 NSP10/NSP16 Methyltransferase in a Complex with SAM Determined by Fixed-Target Serial Crystallography | | Descriptor: | 2'-O-methyltransferase, CHLORIDE ION, Non-structural protein 10, ... | | Authors: | Wilamowski, M, Sherrell, D.A, Minasov, G, Kim, Y, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-26 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6GDY
 
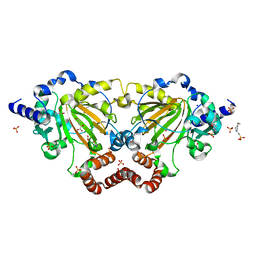 | | Crystal structure of 2OG oxygenase JMJD6 (aa 1-343) in complex with Fe(II) and 2OG | | Descriptor: | 2-OXOGLUTARIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, ... | | Authors: | Islam, M.S, Schofield, C.J, McDonough, M.A. | | Deposit date: | 2018-04-24 | | Release date: | 2019-04-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Biochemical and structural investigations clarify the substrate selectivity of the 2-oxoglutarate oxygenase JMJD6.
J.Biol.Chem., 294, 2019
|
|
3GI4
 
 | | Crystal structure of protease inhibitor, KB60 in complex with wild type HIV-1 protease | | Descriptor: | 5S)-N-[(1S,2R)-3-[(1,3-Benzodioxol-5-ylsulfonyl)(2-methylpropyl)amino]-2-hydroxy-1-(phenylmethyl)propyl]-2-oxo-3-[3-(tr ifluoromethyl)phenyl]-5-oxazolidinecarboxamide, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Nalam, M.N.L, Schiffer, C.A. | | Deposit date: | 2009-03-05 | | Release date: | 2010-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Evaluating the substrate-envelope hypothesis: structural analysis of novel HIV-1 protease inhibitors designed to be robust against drug resistance.
J.Virol., 84, 2010
|
|
5A7R
 
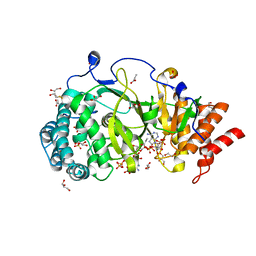 | | Human poly(ADP-ribose) glycohydrolase in complex with synthetic dimeric ADP-ribose | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, POLY(ADP-RIBOSE) GLYCOHYDROLASE, ... | | Authors: | Lambrecht, M.J, Brichacek, M, Barkauskaite, E, Ariza, A, Ahel, I, Hergenrother, P.J. | | Deposit date: | 2015-07-09 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Synthesis of Dimeric Adp-Ribose and its Structure with Human Poly(Adp-Ribose) Glycohydrolase.
J.Am.Chem.Soc., 137, 2015
|
|
