5JF4
 
 | | Crystal structure of type 2 PDF from Streptococcus agalactiae in complex with inhibitor AT019 | | Descriptor: | (3R)-3-{3-[(1-benzofuran-3-yl)methyl]-1,2,4-oxadiazol-5-yl}-4-cyclopentyl-N-hydroxybutanamide, ACETATE ION, IMIDAZOLE, ... | | Authors: | Fieulaine, S, Giglione, C, Meinnel, T. | | Deposit date: | 2016-04-19 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A unique peptide deformylase platform to rationally design and challenge novel active compounds.
Sci Rep, 6, 2016
|
|
5JF5
 
 | | Crystal structure of type 2 PDF from Streptococcus agalactiae in complex with inhibitor AT020 | | Descriptor: | (3R)-3-{3-[(2H-1,3-benzodioxol-5-yl)methyl]-1,2,4-oxadiazol-5-yl}-4-cyclopentyl-N-hydroxybutanamide, ACETATE ION, IMIDAZOLE, ... | | Authors: | Fieulaine, S, Giglione, C, Meinnel, T. | | Deposit date: | 2016-04-19 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A unique peptide deformylase platform to rationally design and challenge novel active compounds.
Sci Rep, 6, 2016
|
|
5JF0
 
 | | Crystal structure of type 2 PDF from Streptococcus agalactiae in complex with tripeptide Met-Ala-Arg | | Descriptor: | ACETATE ION, MET-ALA-ARG, NICKEL (II) ION, ... | | Authors: | Fieulaine, S, Giglione, C, Meinnel, T. | | Deposit date: | 2016-04-19 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A unique peptide deformylase platform to rationally design and challenge novel active compounds.
Sci Rep, 6, 2016
|
|
5JF2
 
 | | Crystal structure of type 2 PDF from Streptococcus agalactiae in complex with inhibitor AT002 | | Descriptor: | (3R)-3-{3-[(4-fluorophenyl)methyl]-1,2,4-oxadiazol-5-yl}-N-hydroxyheptanamide, ACETATE ION, IMIDAZOLE, ... | | Authors: | Fieulaine, S, Giglione, C, Meinnel, T, Hamiche, K. | | Deposit date: | 2016-04-19 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A unique peptide deformylase platform to rationally design and challenge novel active compounds.
Sci Rep, 6, 2016
|
|
1U2T
 
 | | X-Ray structure of the sucrose-phosphatase (SPP) from Synechocystis sp. PCC6803 in complex with sucrose6P | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, sucrose-phosphatase (SPP) | | Authors: | Fieulaine, S, Lunn, J.E, Borel, F, Ferrer, J.-L. | | Deposit date: | 2004-07-20 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of a cyanobacterial sucrose-phosphatase reveals the sugar tongs that release free sucrose in the cell
PLANT CELL, 17, 2005
|
|
6YPT
 
 | |
1KKL
 
 | | L.casei HprK/P in complex with B.subtilis HPr | | Descriptor: | CALCIUM ION, HprK protein, PHOSPHOCARRIER PROTEIN HPR | | Authors: | Fieulaine, S, Morera, S, Poncet, S, Galinier, A, Janin, J, Deutscher, J, Nessler, S. | | Deposit date: | 2001-12-10 | | Release date: | 2002-08-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure of a bifunctional protein kinase in complex with its protein substrate HPr.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
3SVL
 
 | | Structural basis of the improvement of ChrR - a multi-purpose enzyme | | Descriptor: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, protein yieF | | Authors: | Poulain, S, Eswaramoorthy, S, Hienerwadel, R, Bremond, N, Sylvester, M.D, Zhang, Y.B, Van Der Lelie, D, Berthomieu, C, Matin, A.C. | | Deposit date: | 2011-07-12 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of ChrR-A Quinone Reductase with the Capacity to Reduce Chromate.
Plos One, 7, 2012
|
|
1JB1
 
 | | Lactobacillus casei HprK/P Bound to Phosphate | | Descriptor: | HPRK PROTEIN, PHOSPHATE ION | | Authors: | Fieulaine, S, Morera, S, Poncet, S, Monedero, V, Gueguen-Chaignon, V, Galinier, A, Janin, J, Deutscher, J, Nessler, S. | | Deposit date: | 2001-06-01 | | Release date: | 2001-08-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure of HPr kinase: a bacterial protein kinase with a P-loop nucleotide-binding domain.
EMBO J., 20, 2001
|
|
3EKN
 
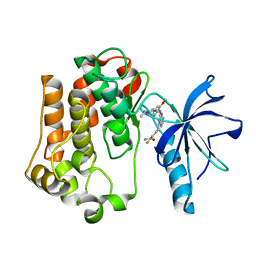 | | Insulin receptor kinase complexed with an inhibitor | | Descriptor: | 2-fluoro-6-{[2-({2-methoxy-4-[4-(1-methylethyl)piperazin-1-yl]phenyl}amino)-7H-pyrrolo[2,3-d]pyrimidin-4-yl]amino}benzamide, Insulin receptor | | Authors: | Chamberlain, S, Atkins, C, Deanda, F, Dumble, M, Gerding, R, Groy, A, Korenchuk, S, Kumar, R, Lei, H, Mook, R, Moorthy, G, Redman, A, Rowland, J, Shewchuk, L. | | Deposit date: | 2008-09-19 | | Release date: | 2008-12-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Optimization of 4,6-bis-anilino-1H-pyrrolo[2,3-d]pyrimidine IGF-1R tyrosine kinase inhibitors towards JNK selectivity.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3EKK
 
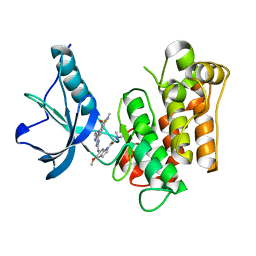 | | Insulin receptor kinase complexed with an inhibitor | | Descriptor: | 2-[(2-{[1-(N,N-dimethylglycyl)-5-methoxy-1H-indol-6-yl]amino}-7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]-6-fluoro-N-methylbenzamide, Insulin receptor | | Authors: | Chamberlain, S, Atkins, C, Deanda, F, Dumble, M, Gerding, R, Groy, A, Korenchuk, S, Kumar, R, Lei, H, Mook, R, Moorthy, G, Redman, A, Rowland, J, Sabbatini, P, Shewchuk, L. | | Deposit date: | 2008-09-19 | | Release date: | 2008-12-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of 4,6-bis-anilino-1H-pyrrolo[2,3-d]pyrimidines: Potent inhibitors of the IGF-1R receptor tyrosine kinase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3ELJ
 
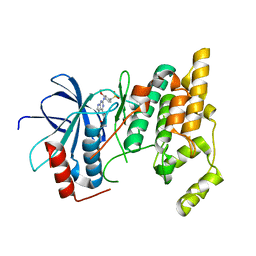 | | Jnk1 complexed with a bis-anilino-pyrrolopyrimidine inhibitor. | | Descriptor: | 2-fluoro-6-{[2-({2-methoxy-4-[(methylsulfonyl)methyl]phenyl}amino)-7H-pyrrolo[2,3-d]pyrimidin-4-yl]amino}benzamide, Mitogen-activated protein kinase 8 | | Authors: | Chamberlain, S, Atkins, C, Deanda, F, Dumble, M, Gerding, R, Groy, A, Korenchuk, S, Kumar, R, Lei, H, Mook, R, Moorthy, G, Redman, A, Rowland, J, Shewchuk, L, Vicentini, G, Mosley, J. | | Deposit date: | 2008-09-22 | | Release date: | 2008-12-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Optimization of 4,6-bis-anilino-1H-pyrrolo[2,3-d]pyrimidine IGF-1R tyrosine kinase inhibitors towards JNK selectivity.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
5MTE
 
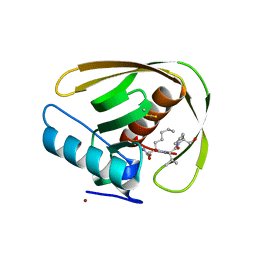 | | Crystal structure of PDF from the Vibrio parahaemolyticus bacteriophage VP16T in complex with actinonin - crystal form II | | Descriptor: | ACTINONIN, NICKEL (II) ION, Putative uncharacterized protein orf60T, ... | | Authors: | Fieulaine, S, Grzela, R, Giglione, C, Meinnel, T. | | Deposit date: | 2017-01-09 | | Release date: | 2017-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Peptide deformylases from Vibrio parahaemolyticus phage and bacteria display similar deformylase activity and inhibitor binding clefts.
Biochim. Biophys. Acta, 1866, 2018
|
|
1U2S
 
 | | X-Ray structure of the sucrose-phosphatase (SPP) from Synechocystis sp. PCC6803 in complex with glucose | | Descriptor: | MAGNESIUM ION, alpha-D-glucopyranose, sucrose-phosphatase | | Authors: | Fieulaine, S, Lunn, J.E, Borel, F, Ferrer, J.-L. | | Deposit date: | 2004-07-20 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of a cyanobacterial sucrose-phosphatase reveals the sugar tongs that release free sucrose in the cell
PLANT CELL, 17, 2005
|
|
3PN2
 
 | |
3PN5
 
 | |
3PN4
 
 | | Crystal structure of Arabidopsis thaliana petide deformylase 1B (AtPDF1B) in complex with actinonin (crystallized in PEG-550-MME) | | Descriptor: | ACTINONIN, Peptide deformylase 1B, chloroplastic, ... | | Authors: | Fieulaine, S, Meinnel, T, Giglione, C. | | Deposit date: | 2010-11-18 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Trapping conformational States along ligand-binding dynamics of Peptide deformylase: the impact of induced fit on enzyme catalysis.
Plos Biol., 9, 2011
|
|
3PN3
 
 | | Crystal structure of Arabidopsis thaliana petide deformylase 1B (AtPDF1B) in complex with inhibitor 21 | | Descriptor: | Peptide deformylase 1B, chloroplastic, ZINC ION, ... | | Authors: | Fieulaine, S, Meinnel, T, Giglione, C. | | Deposit date: | 2010-11-18 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Trapping conformational States along ligand-binding dynamics of Peptide deformylase: the impact of induced fit on enzyme catalysis.
Plos Biol., 9, 2011
|
|
3O3J
 
 | |
3PN6
 
 | |
4JE6
 
 | | Crystal structure of a human-like mitochondrial peptide deformylase | | Descriptor: | Peptide deformylase 1A, chloroplastic/mitochondrial, ZINC ION | | Authors: | Fieulaine, S, Meinnel, T, Giglione, C. | | Deposit date: | 2013-02-26 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Understanding the highly efficient catalysis of prokaryotic peptide deformylases by shedding light on the determinants specifying the low activity of the human counterpart.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4JE8
 
 | | Crystal structure of a human-like mitochondrial peptide deformylase in complex with Met-Ala-Ser | | Descriptor: | Peptide deformylase 1A, chloroplastic/mitochondrial, ZINC ION, ... | | Authors: | Fieulaine, S, Meinnel, T, Giglione, C. | | Deposit date: | 2013-02-26 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Understanding the highly efficient catalysis of prokaryotic peptide deformylases by shedding light on the determinants specifying the low activity of the human counterpart.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5MTD
 
 | | Crystal structure of PDF from the Vibrio parahaemolyticus bacteriophage VP16T - crystal form II | | Descriptor: | NICKEL (II) ION, Putative uncharacterized protein orf60T, TRIETHYLENE GLYCOL, ... | | Authors: | Fieulaine, S, Grzela, R, Giglione, C, Meinnel, T. | | Deposit date: | 2017-01-09 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The C-terminal residue of phage Vp16 PDF, the smallest peptide deformylase, acts as an offset element locking the active conformation.
Sci Rep, 7, 2017
|
|
5MTC
 
 | | Crystal structure of PDF from the Vibrio parahaemolyticus bacteriophage VP16T - crystal form I | | Descriptor: | NICKEL (II) ION, Putative uncharacterized protein orf60T, ZINC ION | | Authors: | Fieulaine, S, Grzela, R, Giglione, C, Meinnel, T. | | Deposit date: | 2017-01-09 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The C-terminal residue of phage Vp16 PDF, the smallest peptide deformylase, acts as an offset element locking the active conformation.
Sci Rep, 7, 2017
|
|
4JE7
 
 | | Crystal structure of a human-like mitochondrial peptide deformylase in complex with actinonin | | Descriptor: | ACTINONIN, Peptide deformylase 1A, chloroplastic/mitochondrial, ... | | Authors: | Fieulaine, S, Meinnel, T, Giglione, C. | | Deposit date: | 2013-02-26 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Understanding the highly efficient catalysis of prokaryotic peptide deformylases by shedding light on the determinants specifying the low activity of the human counterpart.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
