1WBX
 
 | | CRYSTAL STRUCTURES OF MURINE MHC CLASS I H-2 Db AND Kb MOLECULES IN COMPLEX WITH CTL EPITOPES FROM INFLUENZA A VIRUS: IMPLICATIONS FOR TCR REPERTOIRE SELECTION AND IMMUNODOMINANCE | | Descriptor: | BETA-2MICROGLOBULIN, H-2 CLASS I HISTOCOMPATIBILITY ANTIGEN, D-B ALPHA CHAIN, ... | | Authors: | Meijers, R, Lai, C, Yang, Y, Liu, J, Zhong, W, Wang, J, Reinherz, E.L. | | Deposit date: | 2004-11-05 | | Release date: | 2005-01-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Murine Mhc Class I H-2 D(B) and K(B) Molecules in Complex with Ctl Epitopes from Influenza a Virus: Implications for Tcr Repertoire Selection and Immunodominance
J.Mol.Biol., 345, 2005
|
|
1WBY
 
 | | CRYSTAL STRUCTURES OF MURINE MHC CLASS I H-2 Db AND Kb MOLECULES IN COMPLEX WITH CTL EPITOPES FROM INFLUENZA A VIRUS: IMPLICATIONS FOR TCR REPERTOIRE SELECTION AND IMMUNODOMINANCE | | Descriptor: | BETA-2MICROGLOBULIN, H-2 CLASS I HISTOCOMPATIBILITY ANTIGEN, D-B ALPHA CHAIN, ... | | Authors: | Meijers, R, Lai, C, Yang, Y, Liu, J, Zhong, W, Wang, J, Reinherz, E.L. | | Deposit date: | 2004-11-05 | | Release date: | 2005-01-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Murine Mhc Class I H-2 D(B) and K(B) Molecules in Complex with Ctl Epitopes from Influenza a Virus: Implications for Tcr Repertoire Selection and Immunodominance
J.Mol.Biol., 345, 2005
|
|
4OYR
 
 | | Competition of the small inhibitor PT91 with large fatty acyl substrate of the Mycobacterium tuberculosis enoyl-ACP reductase InhA by induced substrate-binding loop refolding | | Descriptor: | 2-(2-chloranylphenoxy)-5-hexyl-phenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Li, H.J, Pan, P, Lai, C.T, Liu, N, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-02-13 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2995 Å) | | Cite: | A Structural and Energetic Model for the Slow-Onset Inhibition of the Mycobacterium tuberculosis Enoyl-ACP Reductase InhA.
Acs Chem.Biol., 9, 2014
|
|
4OXK
 
 | | Multiple binding modes of inhibitor PT155 to the Mycobacterium tuberculosis enoyl-ACP reductase InhA within a tetramer | | Descriptor: | 3,6,9,12,15-pentaoxaoctadecan-17-amine, 5-(4-amino-2-methylphenoxy)-2-hexyl-4-hydroxy-1-methylpyridinium, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Li, H.J, Pan, P, Lai, C.T, Liu, N, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-02-05 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8429 Å) | | Cite: | A Structural and Energetic Model for the Slow-Onset Inhibition of the Mycobacterium tuberculosis Enoyl-ACP Reductase InhA.
Acs Chem.Biol., 9, 2014
|
|
4OXY
 
 | | Substrate-binding loop movement with inhibitor PT10 in the tetrameric Mycobacterium tuberculosis enoyl-ACP reductase InhA | | Descriptor: | 5-hexyl-2-(2-nitrophenoxy)phenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Li, H.J, Sullivan, T.J, Pan, P, Lai, C.T, Liu, N, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-02-09 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3501 Å) | | Cite: | A Structural and Energetic Model for the Slow-Onset Inhibition of the Mycobacterium tuberculosis Enoyl-ACP Reductase InhA.
Acs Chem.Biol., 9, 2014
|
|
7CF7
 
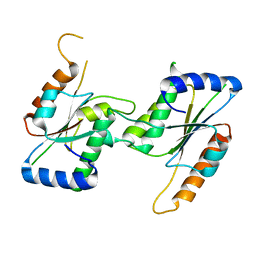 | | Crystal Structure of YbeA CP74 W72F | | Descriptor: | Ribosomal RNA large subunit methyltransferase H,Ribosomal RNA large subunit methyltransferase H | | Authors: | Liu, C.Y, Lai, C.H, Hsu, S.T.D, Lyu, P.C. | | Deposit date: | 2020-06-24 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6178 Å) | | Cite: | Crystal Structure of YbeA CP74 W72F
To Be Published
|
|
7CFY
 
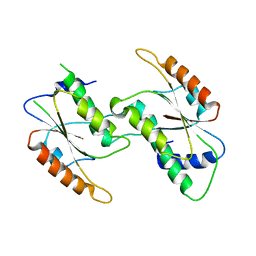 | | Crystal Structure of YbeA CP74 W48F | | Descriptor: | Ribosomal RNA large subunit methyltransferase H,Ribosomal RNA large subunit methyltransferase H | | Authors: | Liu, C.Y, Lai, C.H, Hsu, S.T.D, Lyu, P.C. | | Deposit date: | 2020-06-29 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4051 Å) | | Cite: | Crystal Structure of YbeA CP74 W48F
To Be Published
|
|
7CIU
 
 | |
7CIV
 
 | |
4OXN
 
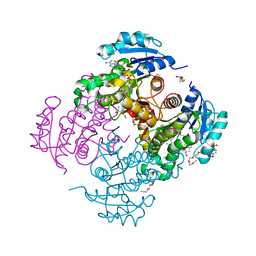 | | Substrate-like binding mode of inhibitor PT155 to the Mycobacterium tuberculosis enoyl-ACP reductase InhA | | Descriptor: | 3,6,9,12,15-pentaoxaoctadecan-17-amine, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-(4-amino-2-methylphenoxy)-2-hexyl-4-hydroxy-1-methylpyridinium, ... | | Authors: | Li, H.J, Pan, P, Lai, C.T, Liu, N, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-02-05 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2926 Å) | | Cite: | A Structural and Energetic Model for the Slow-Onset Inhibition of the Mycobacterium tuberculosis Enoyl-ACP Reductase InhA.
Acs Chem.Biol., 9, 2014
|
|
2I3Z
 
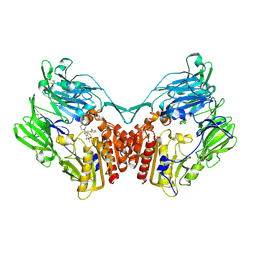 | | rat DPP-IV with xanthine mimetic inhibitor #7 | | Descriptor: | 2-[(3S)-3-AMINOPIPERIDIN-1-YL]-1-(2-CYANOBENZYL)-5-METHYL-4,6-DIOXO-3,4,5,6-TETRAHYDROPYRROLO[3,4-D]IMIDAZOL-1-IUM, Dipeptidyl peptidase 4 (Dipeptidyl peptidase IV) (DPP IV) | | Authors: | Kurukulasuriya, R, Rohde, J.J, Szczepankiewicz, B.G, Basha, F, Lai, C, Winn, M, Stewart, K.D, Longenecker, K.L, Lubben, T.W, Ballaron, S.J, Sham, H.L, VonGeldern, T.W. | | Deposit date: | 2006-08-21 | | Release date: | 2006-12-12 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Xanthine mimetics as potent dipeptidyl peptidase IV inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2OAG
 
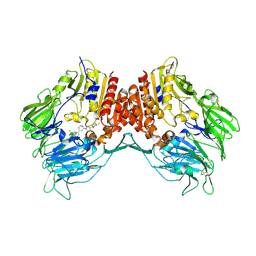 | | Crystal structure of human dipeptidyl peptidase IV (DPPIV) with pyrrolidine-constrained phenethylamine 29g | | Descriptor: | (3R,4S)-1-{6-[3-(METHYLSULFONYL)PHENYL]PYRIMIDIN-4-YL}-4-(2,4,5-TRIFLUOROPHENYL)PYRROLIDIN-3-AMINE, Dipeptidyl peptidase 4 | | Authors: | Backes, B.J, Longenecker, K.L, Hamilton, G.L, Stewart, K.D, Lai, C, Kopecka, H. | | Deposit date: | 2006-12-15 | | Release date: | 2007-09-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Pyrrolidine-constrained phenethylamines: The design of potent, selective, and pharmacologically efficacious dipeptidyl peptidase IV (DPP4) inhibitors from a lead-like screening hit.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
7Y7L
 
 | | Solution structure of zinc finger domain 2 of human ZFAND1 | | Descriptor: | AN1-type zinc finger protein 1, ZINC ION | | Authors: | Fang, P.J, Lai, C.H, Ko, K.T, Chang, C.F, Hsu, S.T.D. | | Deposit date: | 2022-06-22 | | Release date: | 2023-06-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of p97 recognition by human ZFAND1
To Be Published
|
|
7YAB
 
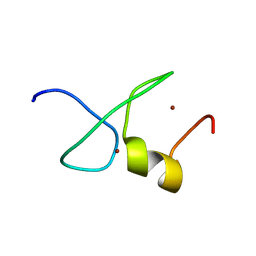 | | Solution structure of zinc finger domain 1 of human ZFAND1 | | Descriptor: | AN1-type zinc finger protein 1, ZINC ION | | Authors: | Fang, P.J, Lai, C.H, Ko, K.T, Chang, C.F, Hsu, S.T.D. | | Deposit date: | 2022-06-27 | | Release date: | 2023-06-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of p97 recognition by human ZFAND1
To Be Published
|
|
2OQI
 
 | | Human Dipeptidyl Peptidase IV (DPP4) with Piperidinone-constrained phenethylamine | | Descriptor: | (4R,5R)-5-AMINO-1-[2-(1,3-BENZODIOXOL-5-YL)ETHYL]-4-(2,4,5-TRIFLUOROPHENYL)PIPERIDIN-2-ONE, Dipeptidyl peptidase 4 (Dipeptidyl peptidase IV) (DPP IV) (T-cell activation antigen CD26) (TP103) (Adenosine deaminase complexing protein 2) (ADABP) | | Authors: | Pei, Z, Li, X, von Geldern, T.W, Longenecker, K.L, Pireh, D, Stewart, K.D, Backes, B.J, Lai, C, Lubben, T.H, Ballaron, S.J, Beno, D.W, Kempf-Grote, A.J, Sham, H.L, Trevillyan, J.M. | | Deposit date: | 2007-01-31 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and Structure-Activity Relationships of Piperidinone- and Piperidine-Constrained Phenethylamines as Novel, Potent, and Selective Dipeptidyl Peptidase IV Inhibitors.
J.Med.Chem., 50, 2007
|
|
2LW3
 
 | | Solution structure of the soluble domain of MmpS4 from Mycobacterium tuberculosis | | Descriptor: | Putative membrane protein mmpS4 | | Authors: | Xi, Z, Sun, P, Wang, W, Lai, C, Wu, F, Tian, C. | | Deposit date: | 2012-07-19 | | Release date: | 2013-03-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Discovery of a Siderophore Export System Essential for Virulence of Mycobacterium tuberculosis
Plos Pathog., 9, 2013
|
|
