6LHF
 
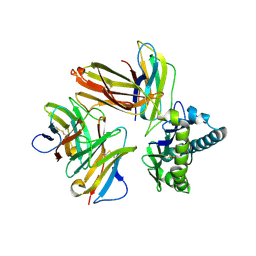 | | Crystal structure of chicken cCD8aa/pBF2*15:01 | | 分子名称: | Beta-2-microglobulin, CD8 alpha chain, MHC class I, ... | | 著者 | Liu, Y.J, Xia, C. | | 登録日 | 2019-12-08 | | 公開日 | 2020-12-09 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | The Combination of CD8 alpha alpha and Peptide-MHC-I in a Face-to-Face Mode Promotes Chicken gamma delta T Cells Response.
Front Immunol, 11, 2020
|
|
6LHG
 
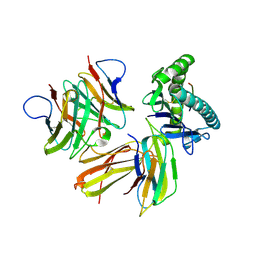 | | Crystal structure of chicken cCD8aa/pBF2*04:01 | | 分子名称: | Beta-2-microglobulin, CD8 alpha chain, IE8 peptide, ... | | 著者 | Liu, Y.J, Xia, C. | | 登録日 | 2019-12-08 | | 公開日 | 2020-12-09 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The Combination of CD8 alpha alpha and Peptide-MHC-I in a Face-to-Face Mode Promotes Chicken gamma delta T Cells Response.
Front Immunol, 11, 2020
|
|
3SGU
 
 | |
4GZN
 
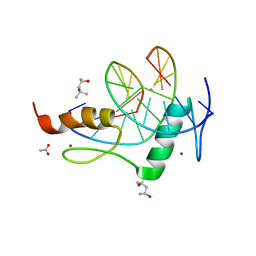 | | Mouse ZFP57 zinc fingers in complex with methylated DNA | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | 著者 | Liu, Y, Zhang, X, Cheng, X. | | 登録日 | 2012-09-06 | | 公開日 | 2012-10-17 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (0.99 Å) | | 主引用文献 | An atomic model of Zfp57 recognition of CpG methylation within a specific DNA sequence.
Genes Dev., 26, 2012
|
|
4OR6
 
 | |
4OR4
 
 | |
3SW6
 
 | |
7BWN
 
 | | Crystal Structure of a Designed Protein Heterocatenane | | 分子名称: | Cellular tumor antigen p53, Chimera of Green fluorescent protein and p53dim | | 著者 | Liu, Y.J, Duan, Z.L, Fang, J, Zhang, F, Xiao, J.Y, Zhang, W.B. | | 登録日 | 2020-04-15 | | 公開日 | 2020-06-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.396 Å) | | 主引用文献 | Cellular Synthesis and X-ray Crystal Structure of a Designed Protein Heterocatenane.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
2L8O
 
 | | Solution structure of Chr148 from Cytophaga hutchinsonii, Northeast Structural Genomics Consortium Target Chr148 | | 分子名称: | Activator of Hsp90 ATPase homologue 1-like C-terminal domain-containing protein | | 著者 | Liu, Y, Lee, D, Ciccosanti, C, Nair, L, Rost, B, Acton, T, Xiao, R, Everett, J, Montelione, G, Prestegard, J, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2011-01-21 | | 公開日 | 2011-03-02 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of Chr148 from Cytophaga hutchinsonii. Northeast Structural Genomics Consortium Target Chr148
To be Published
|
|
3S9Y
 
 | | Crystal Structure of P. falciparum orotidine 5'-monophosphate decarboxylase complexed with 5-fluoro-6-amino-UMP in space group P21, produced from 5-fluoro-6-azido-UMP | | 分子名称: | 6-amino-5-fluorouridine 5'-(dihydrogen phosphate), DI(HYDROXYETHYL)ETHER, HEXAETHYLENE GLYCOL, ... | | 著者 | Liu, Y, Kotra, L.P, Pai, E.F. | | 登録日 | 2011-06-02 | | 公開日 | 2012-04-18 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure-activity relationships of orotidine-5'-monophosphate decarboxylase inhibitors as anticancer agents.
J.Med.Chem., 52, 2009
|
|
3TEJ
 
 | |
3THQ
 
 | |
4O62
 
 | | CW-type zinc finger of ZCWPW2 in complex with the amino terminus of histone H3 | | 分子名称: | Histone H3.3, UNKNOWN ATOM OR ION, ZINC ION, ... | | 著者 | Liu, Y, Tempel, W, Dong, A, Loppnau, P, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2013-12-20 | | 公開日 | 2014-03-26 | | 最終更新日 | 2016-06-08 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Family-wide Characterization of Histone Binding Abilities of Human CW Domain-containing Proteins.
J.Biol.Chem., 291, 2016
|
|
3VAC
 
 | |
3VFO
 
 | | crystal structure of HLA B*3508 LPEP157A, HLA mutant Ala157 | | 分子名称: | Beta-2-microglobulin, LPEP peptide from EBV, LPEPLPQGQLTAY, ... | | 著者 | Liu, Y.C, Rossjohn, J, Gras, S. | | 登録日 | 2012-01-10 | | 公開日 | 2012-03-07 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The Energetic Basis Underpinning T-cell Receptor Recognition of a Super-bulged Peptide Bound to a Major Histocompatibility Complex Class I Molecule.
J.Biol.Chem., 287, 2012
|
|
5YVX
 
 | |
3VFM
 
 | | crystal structure of HLA B*3508 LPEP155A, HLA mutant Ala155 | | 分子名称: | Beta-2-microglobulin, LPEP peptide from EBV, LPEPLPQGQLTAY, ... | | 著者 | Liu, Y.C, Rossjohn, J, Gras, S. | | 登録日 | 2012-01-10 | | 公開日 | 2012-03-07 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The Energetic Basis Underpinning T-cell Receptor Recognition of a Super-bulged Peptide Bound to a Major Histocompatibility Complex Class I Molecule.
J.Biol.Chem., 287, 2012
|
|
3VFV
 
 | | crystal structure of HLA B*3508 LPEP-P9Ala, peptide mutant P9-ala | | 分子名称: | Beta-2-microglobulin, LPEP peptide from EBV, P9A, ... | | 著者 | Liu, Y.C, Rossjohn, J, Gras, S. | | 登録日 | 2012-01-10 | | 公開日 | 2012-02-22 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | The Energetic Basis Underpinning T-cell Receptor Recognition of a Super-bulged Peptide Bound to a Major Histocompatibility Complex Class I Molecule.
J.Biol.Chem., 287, 2012
|
|
3VFW
 
 | | crystal structure of HLA B*3508 LPEP-P10Ala, peptide mutant P10-ala | | 分子名称: | Beta-2-microglobulin, LPEP peptide from EBV, P10A, ... | | 著者 | Liu, Y.C, Rossjohn, J, Gras, S. | | 登録日 | 2012-01-10 | | 公開日 | 2012-02-22 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The Energetic Basis Underpinning T-cell Receptor Recognition of a Super-bulged Peptide Bound to a Major Histocompatibility Complex Class I Molecule.
J.Biol.Chem., 287, 2012
|
|
3VFT
 
 | | crystal structure of HLA B*3508LPEP-P6Ala, peptide mutant P6-ala | | 分子名称: | Beta-2-microglobulin, LPEP peptide from EBV, P6A, ... | | 著者 | Liu, Y.C, Rossjohn, J, Gras, S. | | 登録日 | 2012-01-10 | | 公開日 | 2012-02-22 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.947 Å) | | 主引用文献 | The Energetic Basis Underpinning T-cell Receptor Recognition of a Super-bulged Peptide Bound to a Major Histocompatibility Complex Class I Molecule.
J.Biol.Chem., 287, 2012
|
|
3VFS
 
 | | crystal structure of HLA B*3508LPEP-P5Ala , peptide mutant P5-ala | | 分子名称: | Beta-2-microglobulin, LPEP peptide from EBV, P5A, ... | | 著者 | Liu, Y.C, Rossjohn, J, Gras, S. | | 登録日 | 2012-01-10 | | 公開日 | 2012-02-22 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The Energetic Basis Underpinning T-cell Receptor Recognition of a Super-bulged Peptide Bound to a Major Histocompatibility Complex Class I Molecule.
J.Biol.Chem., 287, 2012
|
|
3VFU
 
 | | crystal structure of HLA B*3508 LPEP-P7Ala, peptide mutant P7-ala | | 分子名称: | Beta-2-microglobulin, LPEP peptide from EBV, P7A, ... | | 著者 | Liu, Y.C, Rossjohn, J, Gras, S. | | 登録日 | 2012-01-10 | | 公開日 | 2012-02-22 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | The Energetic Basis Underpinning T-cell Receptor Recognition of a Super-bulged Peptide Bound to a Major Histocompatibility Complex Class I Molecule.
J.Biol.Chem., 287, 2012
|
|
3VFP
 
 | | crystal structure of HLA B*3508 LPEP158G, HLA mutant Gly158 | | 分子名称: | ACETATE ION, Beta-2-microglobulin, LPEP peptide from EBV, ... | | 著者 | Liu, Y.C, Rossjohn, J, Gras, S. | | 登録日 | 2012-01-10 | | 公開日 | 2012-03-07 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The Energetic Basis Underpinning T-cell Receptor Recognition of a Super-bulged Peptide Bound to a Major Histocompatibility Complex Class I Molecule.
J.Biol.Chem., 287, 2012
|
|
3VFR
 
 | | crystal structure of HLA B*3508LPEP-P4Ala, peptide mutant P4-ala | | 分子名称: | Beta-2-microglobulin, LPEP peptide from EBV, P4A, ... | | 著者 | Liu, Y.C, Rossjohn, J, Gras, S. | | 登録日 | 2012-01-10 | | 公開日 | 2012-03-07 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The Energetic Basis Underpinning T-cell Receptor Recognition of a Super-bulged Peptide Bound to a Major Histocompatibility Complex Class I Molecule.
J.Biol.Chem., 287, 2012
|
|
2KSQ
 
 | | The myristoylated yeast ARF1 in a GTP and bicelle bound conformation | | 分子名称: | ADP-ribosylation factor 1, GUANOSINE-5'-TRIPHOSPHATE, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | 著者 | Liu, Y, Kahn, R, Prestegard, J. | | 登録日 | 2010-01-12 | | 公開日 | 2010-07-07 | | 最終更新日 | 2011-07-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Dynamic structure of membrane-anchored Arf*GTP.
Nat.Struct.Mol.Biol., 17, 2010
|
|
