3TO2
 
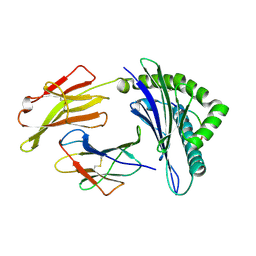 | | Structure of HLA-A*0201 complexed with peptide Md3-C9 derived from a clustering region of restricted cytotoxic T lymphocyte epitope from SARS-CoV M protein | | 分子名称: | Beta-2-microglobulin, MHC class I antigen, Md3-C9 peptide derived from Membrane glycoprotein | | 著者 | Liu, J, Qi, J, Gao, F, Yan, J, Gao, G.F. | | 登録日 | 2011-09-03 | | 公開日 | 2012-08-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Functional and Structural Definition of a Clustering Region of HLA-A2-restricted Cytotoxic T Lymphocyte Epitopes
Sci.Technology Rev., 29, 2011
|
|
4IDT
 
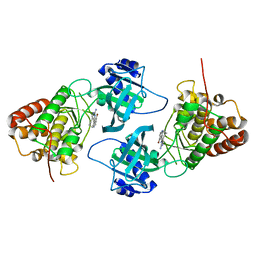 | | Crystal Structure of NIK with 11-bromo-5,6,7,8-tetrahydropyrimido[4',5':3,4]cyclohepta[1,2-b]indol-2-amine (T28) | | 分子名称: | 11-bromo-5,6,7,8-tetrahydropyrimido[4',5':3,4]cyclohepta[1,2-b]indol-2-amine, Mitogen-activated protein kinase kinase kinase 14 | | 著者 | Liu, J, Sudom, A, Wang, Z. | | 登録日 | 2012-12-13 | | 公開日 | 2013-04-17 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Inhibiting NF-KB-inducing kinase (NIK): Discovery, structure-based design, synthesis, structure activity relationship, and co-crystal structures
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4OED
 
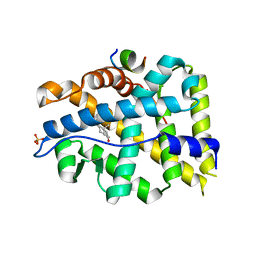 | | Crystal structure of AR-LBD bound with co-regulator peptide | | 分子名称: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, Protein BUD31 homolog, ... | | 著者 | Liu, J.S, Hsu, C.L, Wu, W.G. | | 登録日 | 2014-01-13 | | 公開日 | 2014-08-20 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
4OIL
 
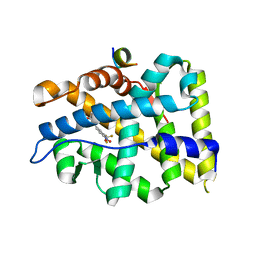 | | Crystal structure of T877A-AR-LBD bound with co-regulator peptide | | 分子名称: | Androgen receptor, HYDROXYFLUTAMIDE, co-regulator peptide | | 著者 | Liu, J.S, Hsu, C.L, Wu, W.G. | | 登録日 | 2014-01-19 | | 公開日 | 2014-08-20 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
4OKW
 
 | | Crystal structure of W741L-AR-LBD bound with co-regulator peptide | | 分子名称: | Androgen receptor, R-BICALUTAMIDE, co-regulator peptide | | 著者 | Liu, J.S, Hsu, C.L, Wu, W.G. | | 登録日 | 2014-01-23 | | 公開日 | 2014-08-20 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
3OMN
 
 | | Catalytic core subunits (I and II) of cytochrome C oxidase from Rhodobacter sphaeroides with D132A mutation in the reduced state | | 分子名称: | (2S,3R)-heptane-1,2,3-triol, CADMIUM ION, CALCIUM ION, ... | | 著者 | Liu, J, Qin, L, Ferguson-Miller, S. | | 登録日 | 2010-08-27 | | 公開日 | 2011-02-02 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystallographic and online spectral evidence for role of conformational change and conserved water in cytochrome oxidase proton pump.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
8I0H
 
 | |
4OK1
 
 | | Crystal structure of W741L-AR-LBD bound with co-regulator peptide | | 分子名称: | Androgen receptor, R-BICALUTAMIDE, co-regulator peptide | | 著者 | Liu, J.S, Hsu, C.L, Wu, W.G. | | 登録日 | 2014-01-21 | | 公開日 | 2014-08-20 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
4OH5
 
 | | Crystal structure of T877A-AR-LBD bound with co-regulator peptide | | 分子名称: | Androgen receptor, HYDROXYFLUTAMIDE, SULFATE ION, ... | | 著者 | Liu, J.S, Hsu, C.L, Wu, W.G. | | 登録日 | 2014-01-17 | | 公開日 | 2014-08-20 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
4OHA
 
 | | Crystal structure of T877A-AR-LBD bound with co-regulator peptide | | 分子名称: | Androgen receptor, HYDROXYFLUTAMIDE, SULFATE ION, ... | | 著者 | Liu, J.S, Hsu, C.L, Wu, W.G. | | 登録日 | 2014-01-17 | | 公開日 | 2014-08-20 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
4OJ9
 
 | | Crystal structure of T877A-AR-LBD bound with co-regulator peptide | | 分子名称: | Androgen receptor, GLYCEROL, HYDROXYFLUTAMIDE, ... | | 著者 | Liu, J.S, Hsu, C.L, Wu, W.G. | | 登録日 | 2014-01-20 | | 公開日 | 2014-08-20 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.31 Å) | | 主引用文献 | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
3TCP
 
 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC569 | | 分子名称: | 1-[(trans-4-aminocyclohexyl)methyl]-N-butyl-3-(4-fluorophenyl)-1H-pyrazolo[3,4-d]pyrimidin-6-amine, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Liu, J, Yang, C, Simpson, C, DeRyckere, D, Van Deusen, A, Miley, M, Kireev, D.B, Norris-Drouin, J, Sather, S, Hunter, D, Patel, H.S, Janzen, W.P, Machius, M, Johnson, G, Earp, H.S, Graham, D.K, Frye, S, Wang, X. | | 登録日 | 2011-08-09 | | 公開日 | 2012-06-20 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Discovery of Novel Small Molecule Mer Kinase Inhibitors for the Treatment of Pediatric Acute Lymphoblastic Leukemia.
ACS Med Chem Lett, 3, 2012
|
|
4OIU
 
 | | Crystal structure of T877A-AR-LBD bound with co-regulator peptide | | 分子名称: | Androgen receptor, HYDROXYFLUTAMIDE, co-regulator peptide | | 著者 | Liu, J.S, Hsu, C.L, Wu, W.G. | | 登録日 | 2014-01-20 | | 公開日 | 2014-08-20 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.01 Å) | | 主引用文献 | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
4OKT
 
 | | Crystal structure of W741L-AR-LBD bound with co-regulator peptide | | 分子名称: | Androgen receptor, R-BICALUTAMIDE, co-regulator peptide | | 著者 | Liu, J.S, Hsu, C.L, Wu, W.G. | | 登録日 | 2014-01-22 | | 公開日 | 2014-08-20 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
4OLM
 
 | | Crystal structure of W741L-AR-LBD bound with co-regulator peptide | | 分子名称: | Androgen receptor, R-BICALUTAMIDE, co-regulator peptide | | 著者 | Liu, J.S, Hsu, C.L, Wu, W.G. | | 登録日 | 2014-01-24 | | 公開日 | 2014-08-20 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
3OM3
 
 | | Catalytic core subunits (I and II) of cytochrome C oxidase from Rhodobacter sphaeroides with K362M mutation in the reduced state | | 分子名称: | (2S,3R)-heptane-1,2,3-triol, CADMIUM ION, CALCIUM ION, ... | | 著者 | Liu, J, Qin, L, Ferguson-Miller, S. | | 登録日 | 2010-08-26 | | 公開日 | 2011-02-02 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystallographic and online spectral evidence for role of conformational change and conserved water in cytochrome oxidase proton pump.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4OJB
 
 | | Crystal structure of W741L-AR-LBD | | 分子名称: | Androgen receptor, R-BICALUTAMIDE | | 著者 | Liu, J.S, Hsu, C.L, Wu, W.G. | | 登録日 | 2014-01-21 | | 公開日 | 2014-08-20 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
4OKX
 
 | | Crystal structure of W741L-AR-LBD bound with co-regulator peptide | | 分子名称: | Androgen receptor, R-BICALUTAMIDE, SULFATE ION, ... | | 著者 | Liu, J.S, Hsu, C.L, Wu, W.G. | | 登録日 | 2014-01-23 | | 公開日 | 2014-08-20 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
4OEA
 
 | | Crystal structure of AR-LBD | | 分子名称: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor | | 著者 | Liu, J.S, Hsu, C.L, Wu, W.G. | | 登録日 | 2014-01-12 | | 公開日 | 2014-08-20 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
4OFR
 
 | | Crystal structure of AR-LBD bound with co-regulator peptide | | 分子名称: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, co-regulator peptide | | 著者 | Liu, J.S, Hsu, C.L, Wu, W.G. | | 登録日 | 2014-01-15 | | 公開日 | 2014-08-20 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
4OFU
 
 | | Crystal structure of AR-LBD bound with co-regulator peptide | | 分子名称: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, co-regulator peptide | | 著者 | Liu, J.S, Hsu, C.L, Wu, W.G. | | 登録日 | 2014-01-15 | | 公開日 | 2014-08-20 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
5WUW
 
 | |
5WUL
 
 | |
2GUS
 
 | | Conformational Transition between Four- and Five-stranded Phenylalanine Zippers Determined by a Local Packing Interaction | | 分子名称: | Major outer membrane lipoprotein | | 著者 | Liu, J, Zheng, Q, Deng, Y, Kallenbach, N.R, Lu, M. | | 登録日 | 2006-05-01 | | 公開日 | 2006-07-25 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Conformational Transition between Four and Five-stranded Phenylalanine Zippers Determined by a Local Packing Interaction.
J.Mol.Biol., 361, 2006
|
|
4LCY
 
 | |
