8HWD
 
 | | Cryo-EM Structure of D5 ADP form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Primase D5 | | Authors: | Li, Y.N, Zhu, J, Guo, Y.Y, Yan, R.H. | | Deposit date: | 2022-12-29 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insight into the assembly and working mechanism of helicase-primase D5 from Mpox virus.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HWG
 
 | | D5 ATPrS-ADP-ssDNA form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Li, Y.N, Zhu, J, Guo, Y.Y, Yan, R.H. | | Deposit date: | 2022-12-29 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insight into the assembly and working mechanism of helicase-primase D5 from Mpox virus.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HWF
 
 | | Cryo-EM Structure of D5 ADP-ssDNA form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Li, Y.N, Zhu, J, Guo, Y.Y, Yan, R.H. | | Deposit date: | 2022-12-29 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insight into the assembly and working mechanism of helicase-primase D5 from Mpox virus.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HWC
 
 | | Cryo-EM Structure of D5 Apo | | Descriptor: | Primase D5 | | Authors: | Li, Y.N, Zhu, J, Guo, Y.Y, Yan, R.H. | | Deposit date: | 2022-12-29 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insight into the assembly and working mechanism of helicase-primase D5 from Mpox virus.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HWH
 
 | | Cryo-EM Structure of D5 Apo-ssDNA form | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), Primase D5 | | Authors: | Li, Y.N, Zhu, J, Guo, Y.Y, Yan, R.H. | | Deposit date: | 2022-12-29 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insight into the assembly and working mechanism of helicase-primase D5 from Mpox virus.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4N4Y
 
 | |
3N54
 
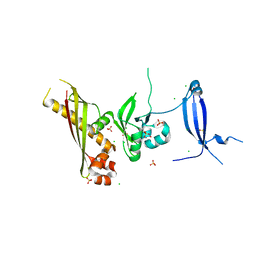 | | Crystal Structure of the GerBC protein | | Descriptor: | CHLORIDE ION, SULFATE ION, Spore germination protein B3 | | Authors: | Li, Y, Setlow, B, Setlow, P, Hao, B. | | Deposit date: | 2010-05-24 | | Release date: | 2010-08-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the GerBC Component of a Bacillus subtilis Spore Germinant Receptor.
J.Mol.Biol., 402, 2010
|
|
8J4I
 
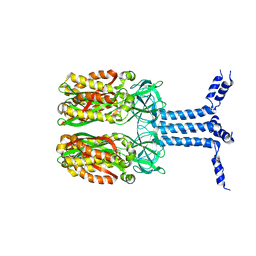 | | Unveiling the Role of Human Prohibitin 2 as a Mitochondrial Calcium Channel in Parkinson's Disease | | Descriptor: | Prohibitin-2 | | Authors: | Li, Y.Y, Fang, Y, Gao, Y.X, Ding, W, Yang, J, Liu, Y, Shen, B, Wang, J.F, Zhou, S. | | Deposit date: | 2023-04-20 | | Release date: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Human Prohibitin 2 is a Mitochondrial Ca2+-Selective Channel
To Be Published
|
|
3EW9
 
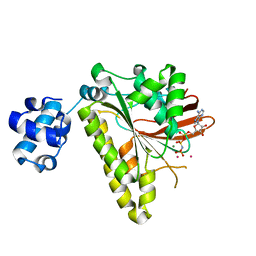 | | RADA recombinase from METHANOCOCCUS MARIPALUDIS in complex with AMPPNP and potassium ions | | Descriptor: | DNA repair and recombination protein radA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, Y, He, Y, Luo, Y. | | Deposit date: | 2008-10-14 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conservation of a conformational switch in RadA recombinase from Methanococcus maripaludis.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
7X8L
 
 | |
3EWA
 
 | | RADA recombinase from METHANOCOCCUS MARIPALUDIS in complex with AMPPNP and ammonium ions | | Descriptor: | DNA repair and recombination protein radA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Li, Y, He, Y, Luo, Y. | | Deposit date: | 2008-10-14 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conservation of a conformational switch in RadA recombinase from Methanococcus maripaludis.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3JBB
 
 | | Characterization of red-shifted phycobiliprotein complexes isolated from the chlorophyll f-containing cyanobacterium Halomicronema hongdechloris | | Descriptor: | PHYCOCYANOBILIN, SULFATE ION, allophycocyanin beta chain, ... | | Authors: | Li, Y, Lin, Y, Garvey, C, Birch, D, Corkery, R.W, Loughlin, P.C, Scheer, H, Willows, R.D, Chen, M. | | Deposit date: | 2015-08-26 | | Release date: | 2015-11-11 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (26 Å) | | Cite: | Characterization of red-shifted phycobilisomes isolated from the chlorophyll f-containing cyanobacterium Halomicronema hongdechloris.
Biochim.Biophys.Acta, 1857, 2015
|
|
3FYH
 
 | | Recombinase in complex with ADP and metatungstate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA repair and recombination protein radA, MAGNESIUM ION, ... | | Authors: | Li, Y, He, Y, Luo, Y. | | Deposit date: | 2009-01-22 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an archaeal Rad51 homologue in complex with a metatungstate inhibitor.
Biochemistry, 48, 2009
|
|
7FAC
 
 | | Crystal Structure of C-terminus of the non-structural protein 2 from SARS coronavirus | | Descriptor: | Non-structural protein 2, ZINC ION | | Authors: | Li, Y.Y, Ren, Z.L, Bao, Z.H, Ming, Z.H, Yan, L.M, Lou, Z.Y, Rao, Z.H. | | Deposit date: | 2021-07-06 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | The Life of SARS-CoV-2 Inside Cells: Replication-Transcription Complex Assembly and Function.
Annu.Rev.Biochem., 91, 2022
|
|
7FA1
 
 | | Crystal Structure of N-terminus of the non-structural protein 2 from SARS coronavirus | | Descriptor: | Non-structural protein 2, ZINC ION | | Authors: | Li, Y.Y, Ren, Z.L, Bao, Z.H, Ming, Z.H, Yan, L.M, Lou, Z.Y, Rao, Z.H. | | Deposit date: | 2021-07-05 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Life of SARS-CoV-2 Inside Cells: Replication-Transcription Complex Assembly and Function.
Annu.Rev.Biochem., 91, 2022
|
|
5YDM
 
 | |
5YDL
 
 | |
5YDA
 
 | |
4OEG
 
 | | Crystal Structure Analysis of FGF2-Disaccharide (S9I2) complex | | Descriptor: | 2-deoxy-3,6-di-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-1-O-methyl-2-O-sulfo-alpha-L-idopyranuronic acid, Fibroblast growth factor 2 | | Authors: | Li, Y.C, Hsiao, C.D. | | Deposit date: | 2014-01-13 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Interactions that influence the binding of synthetic heparan sulfate based disaccharides to fibroblast growth factor-2.
Acs Chem.Biol., 9, 2014
|
|
4OEE
 
 | | Crystal Structure Analysis of FGF2-Disaccharide (S3I2) complex | | Descriptor: | 2-deoxy-3-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-1-O-methyl-2-O-sulfo-alpha-L-idopyranuronic acid, Fibroblast growth factor 2 | | Authors: | Li, Y.C, Hsiao, C.D. | | Deposit date: | 2014-01-13 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Interactions that influence the binding of synthetic heparan sulfate based disaccharides to fibroblast growth factor-2.
Acs Chem.Biol., 9, 2014
|
|
8I9H
 
 | | S-RBD (Omicron XBB.1) in complex with PD of ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of SARS-CoV-2 BA.2-derived subvariants spike in complex with ACE2 receptor.
Cell Discov, 9, 2023
|
|
8I9G
 
 | | S-RBD (Omicron BF.7) in complex with PD of ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of SARS-CoV-2 BA.2-derived subvariants spike in complex with ACE2 receptor.
Cell Discov, 9, 2023
|
|
8I9F
 
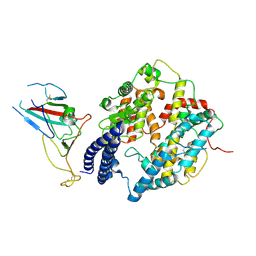 | | S-RBD (Omicron BA.2.75) in complex with PD of ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of SARS-CoV-2 BA.2-derived subvariants spike in complex with ACE2 receptor.
Cell Discov, 9, 2023
|
|
8I9B
 
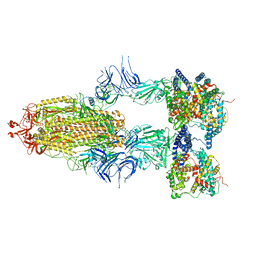 | | S-ECD (Omicron BA.2.75) in complex with PD of ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-07 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of SARS-CoV-2 BA.2-derived subvariants spike in complex with ACE2 receptor.
Cell Discov, 9, 2023
|
|
8I9C
 
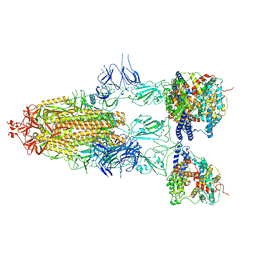 | | S-ECD (Omicron BF.7) in complex with PD of ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-07 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Cryo-EM structures of SARS-CoV-2 BA.2-derived subvariants spike in complex with ACE2 receptor.
Cell Discov, 9, 2023
|
|
