1Y6H
 
 | | Crystal structure of LIPDF | | 分子名称: | FORMIC ACID, GLYCINE, Peptide deformylase, ... | | 著者 | Zhou, Z, Song, X, Li, Y, Gong, W. | | 登録日 | 2004-12-06 | | 公開日 | 2004-12-21 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Unique structural characteristics of peptide deformylase from pathogenic bacterium Leptospira interrogans
J.Mol.Biol., 339, 2004
|
|
1G9I
 
 | | CRYSTAL STRUCTURE OF BETA-TRYSIN COMPLEX IN CYCLOHEXANE | | 分子名称: | BOWMAN-BIRK TYPE TRYPSIN INHIBITOR, CALCIUM ION, SULFATE ION, ... | | 著者 | Zhu, G, Huang, Q, Zhu, Y, Li, Y, Chi, C, Tang, Y. | | 登録日 | 2000-11-24 | | 公開日 | 2000-12-06 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | X-Ray study on an artificial mung bean inhibitor complex with bovine beta-trypsin in neat cyclohexane.
Biochim.Biophys.Acta, 1546, 2001
|
|
2LAS
 
 | | Molecular Determinants of Paralogue-Specific SUMO-SIM Recognition | | 分子名称: | M-IR2_peptide, Small ubiquitin-related modifier 1 | | 著者 | Namanja, A, Li, Y, Su, Y, Wong, S, Lu, J, Colson, L, Wu, C, Li, S, Chen, Y. | | 登録日 | 2011-03-20 | | 公開日 | 2011-12-14 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Insights into High Affinity Small Ubiquitin-like Modifier (SUMO) Recognition by SUMO-interacting Motifs (SIMs) Revealed by a Combination of NMR and Peptide Array Analysis.
J.Biol.Chem., 287, 2012
|
|
9J4I
 
 | | Crystal structure of GH9l Inulin fructotransferases (IFTase) in compex with fruetosyl nystose (GF4) | | 分子名称: | DFA-III-forming inulin fructotransferase, beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-[alpha-D-glucopyranose-(1-2)]beta-D-fructofuranose, beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-[alpha-D-glucopyranose-(1-2)]beta-D-fructofuranose | | 著者 | Chen, G, Wang, Z.X, Yang, Y.Q, Li, Y.G, Zhang, T, Ouyang, S.Y, Zhang, L, Chen, Y, Ruan, X.L, Miao, M. | | 登録日 | 2024-08-09 | | 公開日 | 2024-09-04 | | 最終更新日 | 2024-09-18 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Elucidation of the mechanism underlying the sequential catalysis of inulin by fructotransferase.
Int.J.Biol.Macromol., 277, 2024
|
|
3KYT
 
 | |
3L0J
 
 | | Crystal structure of orphan nuclear receptor RORgamma in complex with natural ligand | | 分子名称: | (3alpha,8alpha,22R)-cholest-5-ene-3,22-diol, Nuclear receptor ROR-gamma, Nuclear receptor coactivator 2 | | 著者 | Martynowski, D, Li, Y. | | 登録日 | 2009-12-10 | | 公開日 | 2010-03-16 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis for hydroxycholesterols as natural ligands of orphan nuclear receptor RORgamma.
Mol.Endocrinol., 24, 2010
|
|
9J4K
 
 | | Crystal structure of GH9l Inulinfructotransferases (IFTase) in complex with GF2 | | 分子名称: | DFA-III-forming inulin fructotransferase, beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | 著者 | Chen, G, Wang, Z.X, Yang, Y.Q, Li, Y.G, Zhang, T, Ouyang, S.Y, Zhang, L, Chen, Y, Ruan, X.L, Miao, M. | | 登録日 | 2024-08-09 | | 公開日 | 2024-09-04 | | 最終更新日 | 2024-09-18 | | 実験手法 | X-RAY DIFFRACTION (2.201 Å) | | 主引用文献 | Elucidation of the mechanism underlying the sequential catalysis of inulin by fructotransferase.
Int.J.Biol.Macromol., 277, 2024
|
|
9J4J
 
 | | Crystal structure of GH9l Inulin fructotransferases(IFTase)incomplex with nystose(F3) | | 分子名称: | DFA-III-forming inulin fructotransferase, beta-D-fructofuranose, beta-D-fructofuranose-(1-1)-beta-D-fructofuranose, ... | | 著者 | Chen, G, Wang, Z.X, Yang, Y.Q, Li, Y.G, Zhang, T, Ouyang, S.Y, Zhang, L, Chen, Y, Ruan, X.L, Miao, M. | | 登録日 | 2024-08-09 | | 公開日 | 2024-09-04 | | 最終更新日 | 2024-09-18 | | 実験手法 | X-RAY DIFFRACTION (2.803 Å) | | 主引用文献 | Elucidation of the mechanism underlying the sequential catalysis of inulin by fructotransferase.
Int.J.Biol.Macromol., 277, 2024
|
|
9J4L
 
 | | Crystal structure of GH9l Inulin fructotransferases (IFTase) | | 分子名称: | DFA-III-forming inulin fructotransferase | | 著者 | Chen, G, Wang, Z.X, Yang, Y.Q, Li, Y.G, Zhang, T, Ouyang, S.Y, Zhang, L, Chen, Y, Ruan, X.L, Miao, M. | | 登録日 | 2024-08-09 | | 公開日 | 2024-09-04 | | 最終更新日 | 2024-09-18 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Elucidation of the mechanism underlying the sequential catalysis of inulin by fructotransferase.
Int.J.Biol.Macromol., 277, 2024
|
|
3F0N
 
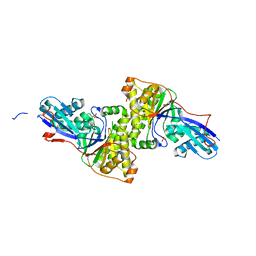 | | Mus Musculus Mevalonate Pyrophosphate Decarboxylase | | 分子名称: | MEVALONATE PYROPHOSPHATE DECARBOXYLASE, PHOSPHATE ION | | 著者 | Walker, J.R, Davis, T, Vesterberg, A, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | 登録日 | 2008-10-25 | | 公開日 | 2008-11-25 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of Mus Musculus Mevalonate Pyrophosphate Decarboxylase
To be Published
|
|
3F83
 
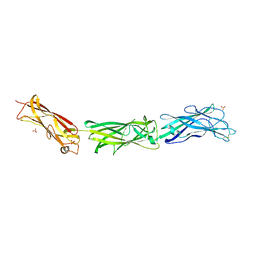 | |
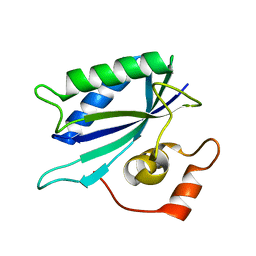
3KUG
 
 | |
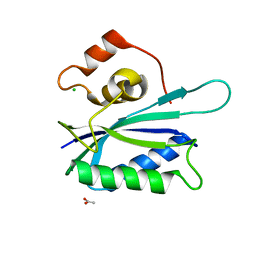
3KUE
 
 | |
3L0L
 
 | |
8EQZ
 
 | | Crystal structure of pregnane X receptor ligand binding domain complexed with T0901317 analog T0-C6 | | 分子名称: | N-[4-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)phenyl]-N-hexylbenzenesulfonamide, Nuclear receptor subfamily 1 group I member 2 | | 著者 | Huber, A.D, Poudel, S, Seetharaman, J, Miller, D.J, Lin, W, Li, Y, Chen, T. | | 登録日 | 2022-10-11 | | 公開日 | 2023-03-15 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | Structure-guided approach to modulate small molecule binding to a promiscuous ligand-activated protein.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3L4H
 
 | | Helical box domain and second WW domain of the human E3 ubiquitin-protein ligase HECW1 | | 分子名称: | ACETIC ACID, E3 ubiquitin-protein ligase HECW1 | | 著者 | Walker, J.R, Qiu, L, Li, Y, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Botchkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | 登録日 | 2009-12-20 | | 公開日 | 2010-05-05 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The tandem helical box and second WW domains of human HECW1
To be Published
|
|
6MB2
 
 | | Cryo-EM structure of the PYD filament of AIM2 | | 分子名称: | Green fluorescent protein, Interferon-inducible protein AIM2 | | 著者 | Lu, A, Li, Y, Wu, H. | | 登録日 | 2018-08-29 | | 公開日 | 2018-09-05 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (5 Å) | | 主引用文献 | Plasticity in PYD assembly revealed by cryo-EM structure of the PYD filament of AIM2.
Cell Discov, 1, 2015
|
|
3KUH
 
 | | Crystal structure of E. coli HPPK(H115A) in complex with AMPCPP and HP | | 分子名称: | 2-AMINO-6-HYDROXYMETHYL-7,8-DIHYDRO-3H-PTERIDIN-4-ONE, 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, ACETATE ION, ... | | 著者 | Blaszczyk, J, Li, Y, Yan, H, Ji, X. | | 登録日 | 2009-11-27 | | 公開日 | 2010-11-24 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Roles of residues E77 and H115 in E. coli HPPK
To be Published
|
|
3F85
 
 | |
3SLI
 
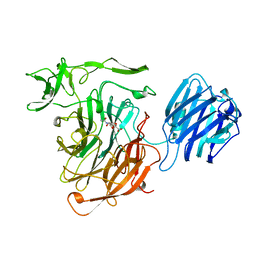 | | LEECH INTRAMOLECULAR TRANS-SIALIDASE COMPLEXED WITH 2,7-ANHYDRO-NEU5AC PREPARED BY SOAKING WITH 3'-SIALYLLACTOSE | | 分子名称: | 2-ACETYLAMINO-7-(1,2-DIHYDROXY-ETHYL)-3-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCTANE-5-CARBOXYLIC ACID, INTRAMOLECULAR TRANS-SIALIDASE | | 著者 | Luo, Y, Li, S.C, Li, Y.T, Luo, M. | | 登録日 | 1998-10-03 | | 公開日 | 1999-04-27 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The 1.8 A structures of leech intramolecular trans-sialidase complexes: evidence of its enzymatic mechanism.
J.Mol.Biol., 285, 1999
|
|
8SSX
 
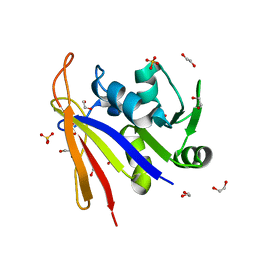 | | Crystal structure of Bacillus anthracis dihydrofolate reductase at 1.65-A resolution | | 分子名称: | 1,2-ETHANEDIOL, Dihydrofolate reductase, SULFATE ION | | 著者 | Shaw, G.X, Li, Y, Wu, Y, Yan, H, Ji, X. | | 登録日 | 2023-05-09 | | 公開日 | 2023-05-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structure of Bacillus anthracis dihydrofolate reductase at 1.65-A resolution
To be published
|
|
3SX0
 
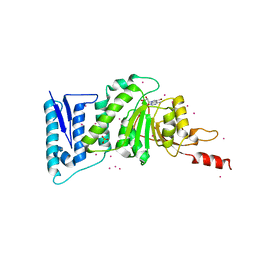 | | Crystal structure of Dot1l in complex with a brominated SAH analog | | 分子名称: | (2S)-2-amino-4-({[(2S,3S,4R,5R)-5-(4-amino-5-bromo-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}sulfanyl)butanoic acid (non-preferred name), Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | 著者 | Yu, W, Tempel, W, Smil, D, Schapira, M, Li, Y, Vedadi, M, Nguyen, K.T, Wernimont, A.K, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Brown, P.J, Structural Genomics Consortium (SGC) | | 登録日 | 2011-07-14 | | 公開日 | 2011-07-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Bromo-deaza-SAH: a potent and selective DOT1L inhibitor.
Bioorg. Med. Chem., 21, 2013
|
|
3HD2
 
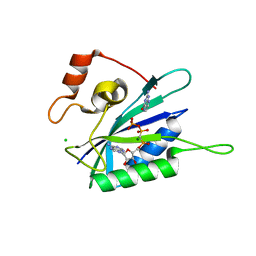 | | Crystal structure of E. coli HPPK(Q50A) in complex with MgAMPCPP and pterin | | 分子名称: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, ACETATE ION, CHLORIDE ION, ... | | 著者 | Blaszczyk, J, Li, Y, Yan, H, Ji, X. | | 登録日 | 2009-05-06 | | 公開日 | 2010-05-19 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Role of loop coupling in enzymatic catalysis and conformational dynamics
To be Published
|
|
2LV4
 
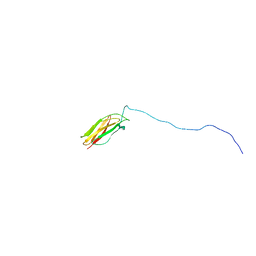 | | ZirS C-terminal Domain | | 分子名称: | Putative outer membrane or exported protein | | 著者 | Prehna, G, Li, Y, Stoynov, N, Okon, M, Vukovic, M, Mcintosh, L.P, Foster, L.J, Finlay, B, Strynadka, N.C.J. | | 登録日 | 2012-06-28 | | 公開日 | 2012-08-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The zinc regulated antivirulence pathway of salmonella is a multiprotein immunoglobulin adhesion system.
J.Biol.Chem., 287, 2012
|
|
3HD1
 
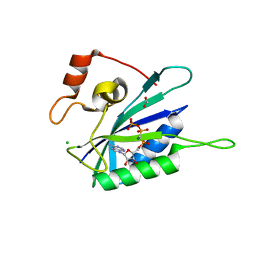 | | Crystal structure of E. coli HPPK(N10A) in complex with MgAMPCPP | | 分子名称: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, ACETATE ION, CHLORIDE ION, ... | | 著者 | Blaszczyk, J, Li, Y, Yan, H, Ji, X. | | 登録日 | 2009-05-06 | | 公開日 | 2010-05-19 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Role of loop coupling in enzymatic catalysis and conformational dynamics
To be Published
|
|
