6MHU
 
 | | Nucleotide-free Cryo-EM Structure of E.coli LptB2FG Transporter | | Descriptor: | (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[(2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-5-[(2~{S},3~{S},4~{R},5~{R},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-4-[(2~{R},3~{S},4~{R},5~{S},6~{R})-6-[(1~{S})-2-[(2~{S},3~{S},4~{S},5~{S},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-1-oxidanyl-ethyl]-3,4-bis(oxidanyl)-5-phosphonooxy-oxan-2-yl]oxy-3-oxidanyl-5-phosphonooxy-oxan-2-yl]oxy-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-dodecanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanyltetradecanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(3~{R})-3-tetradecanoyloxytetradecanoyl]oxy-oxan-2-yl]methoxy]oxan-4-yl]oxy-4,5-bis(oxidanyl)oxane-2-carboxylic acid, Lipopolysaccharide export system ATP-binding protein LptB, Lipopolysaccharide export system permease protein LptF, ... | | Authors: | Orlando, B.J, Li, Y, Liao, M. | | Deposit date: | 2018-09-18 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of lipopolysaccharide extraction by the LptB2FGC complex.
Nature, 567, 2019
|
|
6MI8
 
 | | Cryo-EM Structure of vanadate-trapped E.coli LptB2FGC | | Descriptor: | ADP ORTHOVANADATE, Lipopolysaccharide export system ATP-binding protein LptB, Lipopolysaccharide export system permease protein LptF, ... | | Authors: | Orlando, B.J, Li, Y, Liao, M. | | Deposit date: | 2018-09-19 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of lipopolysaccharide extraction by the LptB2FGC complex.
Nature, 567, 2019
|
|
6MRW
 
 | | 14-meric ClyA pore complex | | Descriptor: | Hemolysin E, chromosomal | | Authors: | Peng, W, de Souza Santos, M, Li, Y, Tomchick, D.R, Orth, K. | | Deposit date: | 2018-10-15 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | High-resolution cryo-EM structures of the E. coli hemolysin ClyA oligomers.
Plos One, 14, 2019
|
|
1EOJ
 
 | | Design of P1' and P3' residues of trivalent thrombin inhibitors and their crystal structures | | Descriptor: | ALPHA THROMBIN, THROMBIN INHIBITOR P798 | | Authors: | Slon-Usakiewicz, J.J, Sivaraman, J, Li, Y, Cygler, M, Konishi, Y. | | Deposit date: | 2000-03-23 | | Release date: | 2000-05-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design of P1' and P3' residues of trivalent thrombin inhibitors and their crystal structures.
Biochemistry, 39, 2000
|
|
6MRT
 
 | | 12-meric ClyA pore complex | | Descriptor: | Hemolysin E, chromosomal | | Authors: | Peng, W, de Souza Santos, M, Li, Y, Tomchick, D.R, Orth, K. | | Deposit date: | 2018-10-15 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | High-resolution cryo-EM structures of the E. coli hemolysin ClyA oligomers.
Plos One, 14, 2019
|
|
8A9W
 
 | | Crystal structure of PulL C-ter domain | | Descriptor: | SULFATE ION, Type II secretion system protein L | | Authors: | Dazzoni, R, Li, Y, Lopez-Castilla, A, Brier, S, Mechaly, A, Cordier, F, Haouz, A, Nilges, M, Francetic, O, Bardiaux, B, Izadi-Pruneyre, N. | | Deposit date: | 2022-06-29 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Structure and dynamic association of an assembly platform subcomplex of the bacterial type II secretion system.
Structure, 31, 2023
|
|
6MHZ
 
 | | Vanadate trapped Cryo-EM Structure of E.coli LptB2FG Transporter | | Descriptor: | ADP ORTHOVANADATE, Lipopolysaccharide export system ATP-binding protein LptB, Lipopolysaccharide export system permease protein LptF, ... | | Authors: | Orlando, B.J, Li, Y, Liao, M. | | Deposit date: | 2018-09-18 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of lipopolysaccharide extraction by the LptB2FGC complex.
Nature, 567, 2019
|
|
6MI7
 
 | | Nucleotide-free Cryo-EM Structure of E.coli LptB2FGC | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, Lipopolysaccharide export system ATP-binding protein LptB, Lipopolysaccharide export system permease protein LptF, ... | | Authors: | Orlando, B.J, Li, Y, Liao, M. | | Deposit date: | 2018-09-19 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis of lipopolysaccharide extraction by the LptB2FGC complex.
Nature, 567, 2019
|
|
6MRU
 
 | | 13-meric ClyA pore complex | | Descriptor: | Hemolysin E, chromosomal | | Authors: | Peng, W, de Souza Santos, M, Li, Y, Tomchick, D.R, Orth, K. | | Deposit date: | 2018-10-15 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | High-resolution cryo-EM structures of the E. coli hemolysin ClyA oligomers.
Plos One, 14, 2019
|
|
1EOL
 
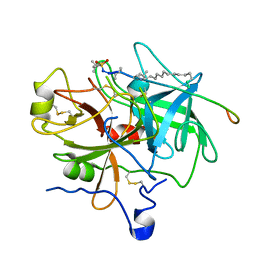 | | Design of P1' and P3' residues of trivalent thrombin inhibitors and their crystal structures | | Descriptor: | ALPHA THROMBIN, THROMBIN INHIBITOR P628 | | Authors: | Slon-Usakiewicz, J.J, Sivaraman, J, Li, Y, Cygler, M, Konishi, Y. | | Deposit date: | 2000-03-23 | | Release date: | 2000-05-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design of P1' and P3' residues of trivalent thrombin inhibitors and their crystal structures.
Biochemistry, 39, 2000
|
|
6JQB
 
 | | The structure of maltooligosaccharide-forming amylase from Pseudomonas saccharophila STB07 with pseudo-maltoheptaose | | Descriptor: | 1,2-ETHANEDIOL, ACARBOSE DERIVED HEPTASACCHARIDE, CALCIUM ION, ... | | Authors: | Li, Z.F, Ban, X.F, Zhang, Z.Q, Li, C.M, Gu, Z.B, Jin, T.C, Li, Y.L, Shang, Y.H. | | Deposit date: | 2019-03-30 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | Structure of maltotetraose-forming amylase from Pseudomonas saccharophila STB07 provides insights into its product specificity.
Int.J.Biol.Macromol., 154, 2020
|
|
4SLI
 
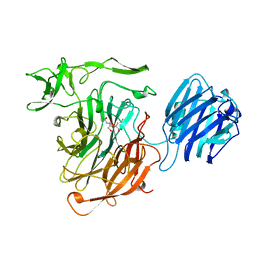 | | LEECH INTRAMOLECULAR TRANS-SIALIDASE COMPLEXED WITH 2-PROPENYL-NEU5AC, AN INACTIVE SUBSTRATE ANALOGUE | | Descriptor: | 2-propenyl-N-acetyl-neuraminic acid, INTRAMOLECULAR TRANS-SIALIDASE | | Authors: | Luo, Y, Li, S.C, Li, Y.T, Luo, M. | | Deposit date: | 1998-10-04 | | Release date: | 1999-05-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.8 A structures of leech intramolecular trans-sialidase complexes: evidence of its enzymatic mechanism.
J.Mol.Biol., 285, 1999
|
|
1EPT
 
 | | REFINED 1.8 ANGSTROMS RESOLUTION CRYSTAL STRUCTURE OF PORCINE EPSILON-TRYPSIN | | Descriptor: | CALCIUM ION, PORCINE E-TRYPSIN | | Authors: | Huang, Q, Wang, Z, Li, Y, Liu, S, Tang, Y. | | Deposit date: | 1994-06-07 | | Release date: | 1995-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined 1.8 A resolution crystal structure of the porcine epsilon-trypsin.
Biochim.Biophys.Acta, 1209, 1994
|
|
1F2E
 
 | | STRUCTURE OF SPHINGOMONAD, GLUTATHIONE S-TRANSFERASE COMPLEXED WITH GLUTATHIONE | | Descriptor: | GLUTATHIONE, GLUTATHIONE S-TRANSFERASE | | Authors: | Nishio, T, Watanabe, T, Patel, A, Wang, Y, Lau, P.C.K, Grochulski, P, Li, Y, Cygler, M. | | Deposit date: | 2000-05-24 | | Release date: | 2000-06-21 | | Last modified: | 2011-12-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Properties of a Sphingomonad and Marine Bacterium Beta-Class Glutathione S-Transferases and Crystal Structure of the Former Complex with Glutathione
To be published
|
|
6WAJ
 
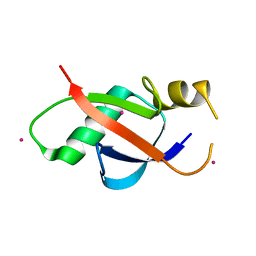 | | Crystal structure of the UBL domain of human NLE1 | | Descriptor: | NLE1, UNKNOWN ATOM OR ION | | Authors: | Halabelian, L, Zeng, H, Li, Y, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-25 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the UBL domain of human NLE1
To be Published
|
|
7MWL
 
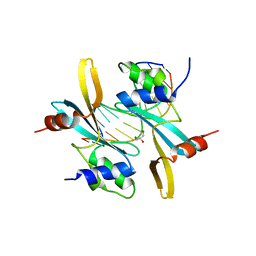 | | The TAM domain of BAZ2A in complex with a 12mer mCG DNA | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, DNA (5'-D(*GP*CP*CP*AP*AP*(5CM)P*GP*TP*TP*GP*GP*C)-3'), GLYCEROL | | Authors: | Liu, K, Dong, A, Li, Y, Loppnau, P, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-17 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The TAM domain of BAZ2A in complex with a 12mer mCG DNA
To Be Published
|
|
6LZ2
 
 | | Crystal structure of a thermostable green fluorescent protein (TGP) with a synthetic nanobody (Sb44) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Cai, H, Yao, H, Li, T, Hutter, C, Tang, Y, Li, Y, Seeger, M, Li, D. | | Deposit date: | 2020-02-17 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | An improved fluorescent tag and its nanobodies for membrane protein expression, stability assay, and purification.
Commun Biol, 3, 2020
|
|
5T0L
 
 | | Crystal structure of Toxoplasma gondii TS-DHFR complexed with NADPH, dUMP, PDDF and 5-(4-(3,4-dichlorophenyl)piperazin-1-yl)pyrimidine-2,4-diamine (TRC-15) | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 5-[4-(3,4-dichlorophenyl)piperazin-1-yl]pyrimidine-2,4-diamine, ... | | Authors: | Thomas, S.B, Chen, Z, Lu, H, Li, Y. | | Deposit date: | 2016-08-16 | | Release date: | 2016-09-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Discovery of Potent and Selective Leads against Toxoplasma gondii Dihydrofolate Reductase via Structure-Based Design.
ACS Med Chem Lett, 7, 2016
|
|
3EFH
 
 | |
3FHQ
 
 | | Structure of endo-beta-N-acetylglucosaminidase A | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, Endo-beta-N-acetylglucosaminidase, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose | | Authors: | Jie, Y, Li, L, Shaw, N, Li, Y, Song, J, Zhang, W, Xia, C, Zhang, R, Joachimiak, A, Zhang, H.-C, Wang, L.-X, Wang, P, Liu, Z.-J. | | Deposit date: | 2008-12-10 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.452 Å) | | Cite: | Structural basis and catalytic mechanism for the dual functional endo-beta-N-acetylglucosaminidase A
Plos One, 4, 2009
|
|
4XCX
 
 | | METHYLTRANSFERASE DOMAIN OF SMALL RNA 2'-O-METHYLTRANSFERASE | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, Small RNA 2'-O-methyltransferase | | Authors: | Walker, J.R, Zeng, H, Dong, A, Li, Y, Wernimont, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-12-18 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal structure of human C1ORF59 in complex with SAH
To be published
|
|
6OEB
 
 | | Crystal structure of HMCES SRAP domain in complex with 3' overhang DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*GP*AP*CP*GP*TP*T)-3'), DNA (5'-D(*GP*TP*CP*TP*GP*G)-3'), ... | | Authors: | Halabelian, L, Ravichandran, M, Li, Y, Zeng, H, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-27 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of HMCES interactions with abasic DNA and multivalent substrate recognition.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6OEA
 
 | | Crystal structure of HMCES SRAP domain in complex with longer 3' overhang DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*GP*AP*CP*GP*TP*TP*GP*TP*T)-3'), DNA (5'-D(*GP*TP*CP*TP*GP*G)-3'), ... | | Authors: | Halabelian, L, Ravichandran, M, Li, Y, Zeng, H, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-27 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of HMCES interactions with abasic DNA and multivalent substrate recognition.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6OE7
 
 | | Crystal structure of HMCES cross-linked to DNA abasic site | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*GP*AP*CP*GP*TP*(DRZ)P*GP*TP*T)-3'), DNA (5'-D(*GP*TP*CP*TP*GP*G)-3'), ... | | Authors: | Halabelian, L, Li, Y, Zeng, H, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-27 | | Release date: | 2019-04-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of HMCES interactions with abasic DNA and multivalent substrate recognition.
Nat.Struct.Mol.Biol., 26, 2019
|
|
4QPN
 
 | | Crystal Structure of Human Methyltransferase-Like Protein 21B | | Descriptor: | Protein-lysine methyltransferase METTL21B, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Tempel, W, Hong, B.S, Seitova, A, He, H, Li, Y, Graslund, S, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-24 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structure of Human Methyltransferase-Like Protein 21B
TO BE PUBLISHED
|
|
