2W31
 
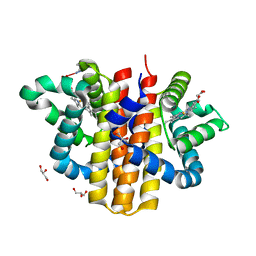 | | globin domain of Geobacter sulfurreducens globin-coupled sensor | | 分子名称: | GLOBIN, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Pesce, A, Thijs, L, Nardini, M, Desmet, F, Sisinni, L, Gourlay, L, Bolli, A, Coletta, M, Van Doorslaer, S, Wan, X, Alam, M, Ascenzi, P, Moens, L, Bolognesi, M, Dewilde, S. | | 登録日 | 2008-11-05 | | 公開日 | 2009-01-13 | | 最終更新日 | 2019-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Hise11 and Hisf8 Provide Bis-Histidyl Heme Hexa-Coordination in the Globin Domain of Geobacter Sulfurreducens Globin-Coupled Sensor.
J.Mol.Biol., 386, 2009
|
|
3ZTF
 
 | | X-ray Structure of the Cyan Fluorescent Protein mTurquoise2 (K206A mutant) | | 分子名称: | GREEN FLUORESCENT PROTEIN | | 著者 | von Stetten, D, Goedhart, J, Noirclerc-Savoye, M, Lelimousin, M, Joosen, L, Hink, M.A, van Weeren, L, Gadella, T.W.J, Royant, A. | | 登録日 | 2011-07-07 | | 公開日 | 2012-03-21 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.31 Å) | | 主引用文献 | Structure-Guided Evolution of Cyan Fluorescent Proteins Towards a Quantum Yield of 93%
Nat.Commun, 3, 2012
|
|
7O7T
 
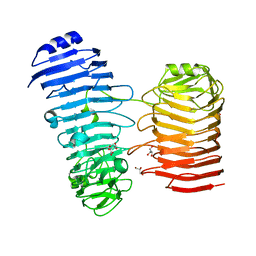 | | Structure of the PL6 family alginate lyase Patl3640 from Pseudoalteromonas atlantica T6c in complex with 4-deoxy-L-erythro-5-hexoseulose uronic acid | | 分子名称: | 1,2-ETHANEDIOL, 4-deoxy-L-erythro-hex-5-ulosuronic acid, GLYCEROL, ... | | 著者 | Ballut, L, Violot, S, Carrique, L, Aghajari, N. | | 登録日 | 2021-04-13 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Exploring molecular determinants of polysaccharide lyase family 6-1 enzyme activity.
Glycobiology, 31, 2021
|
|
7O84
 
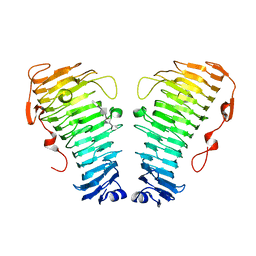 | | Structure of the PL6 family alginate lyase Pedsa0632 from Pseudopedobacter saltans in complex with substrate | | 分子名称: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid, 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid, Alginate lyase | | 著者 | Ballut, L, Violot, S, Carrique, L, Aghajari, N. | | 登録日 | 2021-04-14 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.177 Å) | | 主引用文献 | Exploring molecular determinants of polysaccharide lyase family 6-1 enzyme activity.
Glycobiology, 31, 2021
|
|
7O7A
 
 | |
7O77
 
 | | Structure of the PL6 family alginate lyase Patl3640 from Pseudoalteromonas atlantica T6c | | 分子名称: | GLYCEROL, Poly(Beta-D-mannuronate) lyase | | 著者 | Ballut, L, Violot, S, Carrique, L, Aghajari, N. | | 登録日 | 2021-04-13 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.321 Å) | | 主引用文献 | Exploring molecular determinants of polysaccharide lyase family 6-1 enzyme activity.
Glycobiology, 31, 2021
|
|
7O79
 
 | | Structure of the PL6 family polysaccharide lyase Pedsa3628 from Pseudopedobacter saltans | | 分子名称: | PHOSPHATE ION, Poly(Beta-D-mannuronate) lyase | | 著者 | Ballut, L, Violot, S, Carrique, L, Aghajari, N. | | 登録日 | 2021-04-13 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Exploring molecular determinants of polysaccharide lyase family 6-1 enzyme activity.
Glycobiology, 31, 2021
|
|
7O78
 
 | | Structure of the PL6 family chondroitinase B from Pseudopedobacter saltans, Pedsa3807 | | 分子名称: | Polysaccharide lyase from Pseudopedobacter saltans, Pedsa3807 | | 著者 | Ballut, L, Violot, S, Carrique, L, Aghajari, N. | | 登録日 | 2021-04-13 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Exploring molecular determinants of polysaccharide lyase family 6-1 enzyme activity.
Glycobiology, 31, 2021
|
|
2VR2
 
 | | Human Dihydropyrimidinase | | 分子名称: | CHLORIDE ION, DIHYDROPYRIMIDINASE, ZINC ION | | 著者 | Welin, M, Karlberg, T, Andersson, J, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Herman, M.D, Johansson, I, Kallas, A, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nyman, T, Persson, C, Sagemark, J, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Wikstrom, M, Nordlund, P, Structural Genomics Consortium (SGC) | | 登録日 | 2008-03-25 | | 公開日 | 2008-04-01 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The Crystal Structure of Human Dihydropyrimidinase
To be Published
|
|
3ZJI
 
 | | Tyr(61)B10Ala mutation of M.acetivorans protoglobin in complex with cyanide | | 分子名称: | CYANIDE ION, GLYCEROL, PROTOGLOBIN, ... | | 著者 | Pesce, A, Tilleman, L, Donne, J, Aste, E, Ascenzi, P, Ciaccio, C, Coletta, M, Moens, L, Viappiani, C, Dewilde, S, Bolognesi, M, Nardini, M. | | 登録日 | 2013-01-18 | | 公開日 | 2013-06-26 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure and Haem-Distal Site Plasticity in Methanosarcina Acetivorans Protoglobin.
Plos One, 8, 2013
|
|
2VUX
 
 | | Human ribonucleotide reductase, subunit M2 B | | 分子名称: | FE (III) ION, RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE SUBUNIT M2 B | | 著者 | Welin, M, Moche, M, Andersson, J, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Herman, M.D, Johansson, A, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Nilsson, M.E, Nyman, T, Persson, C, Sagemark, J, Schueler, H, Svensson, L, Thorsell, A.G, Tresaugues, L, van Den Berg, S, Weigelt, J, Wikstrom, M, Nordlund, P, Structural Genomics Consortium (SGC) | | 登録日 | 2008-05-31 | | 公開日 | 2008-07-15 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Human Ribonucleotide Reductase, Subunit M2 B
To be Published
|
|
3ZOL
 
 | | M.acetivorans protoglobin F93Y mutant in complex with cyanide | | 分子名称: | CYANIDE ION, GLYCEROL, PROTOGLOBIN, ... | | 著者 | Tilleman, L, Abbruzzetti, S, Ciaccio, C, De Sanctis, G, Nardini, M, Pesce, A, Desmet, F, Moens, L, Van Doorslaer, S, Bruno, S, Bolognesi, M, Ascenzi, P, Coletta, M, Viappiani, C, Dewilde, S. | | 登録日 | 2013-02-22 | | 公開日 | 2014-03-12 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural Bases for the Regulation of Co Binding in the Archaeal Protoglobin from Methanosarcina Acetivorans.
Plos One, 10, 2015
|
|
3ZOB
 
 | | Solution structure of chicken Engrailed 2 homeodomain | | 分子名称: | HOMEOBOX PROTEIN ENGRAILED-2 | | 著者 | Carlier, L, Balayssac, S, Cantrelle, F.X, Khemtemourian, L, Chassaing, G, Joliot, A, Lequin, O. | | 登録日 | 2013-02-21 | | 公開日 | 2013-08-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Investigation of Homeodomain Membrane Translocation Properties: Insights from the Structure Determination of Engrailed-2 Homeodomain in Aqueous and Membrane-Mimetic Environments.
Biophys.J., 105, 2013
|
|
3ZJH
 
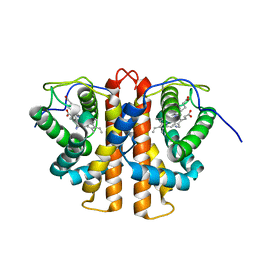 | | Trp(60)B9Ala mutation of M.acetivorans protoglobin in complex with cyanide | | 分子名称: | CYANIDE ION, PROTOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Pesce, A, Tilleman, L, Donne, J, Aste, E, Ascenzi, P, Ciaccio, C, Coletta, M, Moens, L, Viappiani, C, Dewilde, S, Bolognesi, M, Nardini, M. | | 登録日 | 2013-01-18 | | 公開日 | 2013-06-26 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure and Haem-Distal Site Plasticity in Methanosarcina Acetivorans Protoglobin.
Plos One, 8, 2013
|
|
4UVW
 
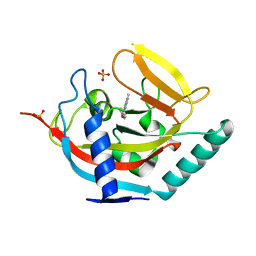 | | Crystal structure of human tankyrase 2 in complex with 4,5-dimethyl-3- phenyl-1,2-dihydroisoquinolin-1-one | | 分子名称: | 4,5-dimethyl-3-phenylisoquinolin-1(2H)-one, SULFATE ION, TANKYRASE-2, ... | | 著者 | Haikarainen, T, Narwal, M, Lehtio, L. | | 登録日 | 2014-08-08 | | 公開日 | 2015-07-29 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Exploration of the Nicotinamide-Binding Site of the Tankyrases, Identifying 3-Arylisoquinolin-1-Ones as Potent and Selective Inhibitors in Vitro.
Bioorg.Med.Chem., 23, 2015
|
|
4UY8
 
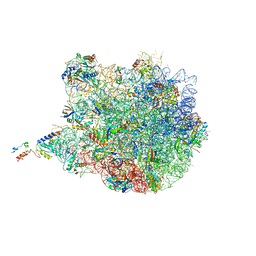 | | Molecular basis for the ribosome functioning as a L-tryptophan sensor - Cryo-EM structure of a TnaC stalled E.coli ribosome | | 分子名称: | 50S RIBOSOMAL PROTEIN L10, 50S RIBOSOMAL PROTEIN L11, 50S RIBOSOMAL PROTEIN L13, ... | | 著者 | Bischoff, L, Berninghausen, O, Beckmann, R. | | 登録日 | 2014-08-29 | | 公開日 | 2014-10-29 | | 最終更新日 | 2018-10-03 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Molecular Basis for the Ribosome Functioning as an L-Tryptophan Sensor.
Cell Rep., 9, 2014
|
|
4V25
 
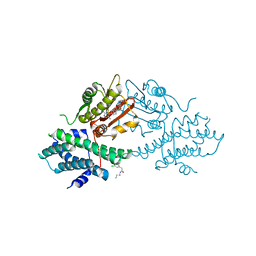 | | VER-246608, a novel pan-isoform ATP competitive inhibitor of pyruvate dehydrogenase kinase, disrupts Warburg metabolism and induces context- dependent cytostasis in cancer cells | | 分子名称: | MAGNESIUM ION, N-(2-AMINOETHYL)-2-{3-CHLORO-4-[(4-ISOPROPYLBENZYL)OXY]PHENYL} ACETAMIDE, N-[4-(2-chloro-5-methylpyrimidin-4-yl)phenyl]-N-(4-{[(difluoroacetyl)amino]methyl}benzyl)-2,4-dihydroxybenzamide, ... | | 著者 | Moore, J.D, Staniszewska, A, Shaw, T, D'Alessandro, J, Davis, B, Surgenor, A, Baker, L, Matassova, N, Murray, J, Macias, A, Brough, P, Wood, M, Mahon, P.C. | | 登録日 | 2014-10-06 | | 公開日 | 2014-12-03 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | VER-246608, a novel pan-isoform ATP competitive inhibitor of pyruvate dehydrogenase kinase, disrupts Warburg metabolism and induces context-dependent cytostasis in cancer cells.
Oncotarget, 5, 2014
|
|
4V4P
 
 | | Crystal structure of 70S ribosome with thrS operator and tRNAs. | | 分子名称: | 16S rRNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Jenner, L, Romby, P, Rees, B, Schulze-Briese, C, Springer, M, Ehresmann, C, Ehresmann, B, Moras, D, Yusupova, G, Yusupov, M. | | 登録日 | 2005-01-19 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (5.5 Å) | | 主引用文献 | Translational operator of mRNA on the ribosome: how repressor proteins exclude ribosome binding.
Science, 308, 2005
|
|
4V87
 
 | | Crystal structure analysis of ribosomal decoding. | | 分子名称: | 16S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | 著者 | Demeshkina, N, Jenner, L, Yusupov, M, Yusupova, G. | | 登録日 | 2011-09-20 | | 公開日 | 2014-07-09 | | 最終更新日 | 2014-12-10 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | A new understanding of the decoding principle on the ribosome.
Nature, 484, 2012
|
|
4UVU
 
 | | Crystal structure of human tankyrase 2 in complex with 1-((4-(5- methyl-1-oxo-1,2-dihydroisoquinolin-3-yl)phenyl)methyl)pyrrolidin-1- ium | | 分子名称: | 5-methyl-3-[4-(pyrrolidin-1-ylmethyl)phenyl]isoquinolin-1(2H)-one, GLYCEROL, SULFATE ION, ... | | 著者 | Haikarainen, T, Narwal, M, Lehtio, L. | | 登録日 | 2014-08-08 | | 公開日 | 2015-07-29 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Exploration of the Nicotinamide-Binding Site of the Tankyrases, Identifying 3-Arylisoquinolin-1-Ones as Potent and Selective Inhibitors in Vitro.
Bioorg.Med.Chem., 23, 2015
|
|
4UVS
 
 | | Crystal structure of human tankyrase 2 in complex with 5-amino-3- pentyl-1,2-dihydroisoquinolin-1-one | | 分子名称: | 5-amino-3-pentylisoquinolin-1(2H)-one, SULFATE ION, TANKYRASE-2, ... | | 著者 | Narwal, M, Haikarainen, T, Lehtio, L. | | 登録日 | 2014-08-08 | | 公開日 | 2015-07-29 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Exploration of the Nicotinamide-Binding Site of the Tankyrases, Identifying 3-Arylisoquinolin-1-Ones as Potent and Selective Inhibitors in Vitro.
Bioorg.Med.Chem., 23, 2015
|
|
4V0I
 
 | | Water Network Determines Selectivity for a Series of Pyrimidone Indoline Amide PI3KBeta Inhibitors over PI3K-Delta | | 分子名称: | 2-[2-(2-METHYL-2,3-DIHYDRO-INDOL-1-YL)-2-OXO-ETHYL]-6-MORPHOLIN-4-YL-3H-PYRIMIDIN-4-ONE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | 著者 | Robinson, D, Bertrand, T, Carry, J.C, Halley, F, Karlsson, A, Mathieu, M, Minoux, H, Perrin, M.A, Robert, B, Schio, L, Sherman, W. | | 登録日 | 2014-09-16 | | 公開日 | 2015-09-30 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.54 Å) | | 主引用文献 | Differential Water Thermodynamics Determine Pi3K-Beta/Delta Selectivity for Solvent-Exposed Ligand Modifications.
J.Chem.Inf.Model., 56, 2016
|
|
4V9R
 
 | | Crystal structure of antibiotic DITYROMYCIN bound to 70S ribosome | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | 著者 | Bulkley, D.P, Brandi, L, Polikanov, Y.S, Fabbretti, A, O'Connor, M, Gualerzi, C.O, Steitz, T.A. | | 登録日 | 2013-12-05 | | 公開日 | 2014-07-09 | | 最終更新日 | 2014-12-10 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The antibiotics dityromycin and GE82832 bind protein S12 and block EF-G-catalyzed translocation.
Cell Rep, 6, 2014
|
|
11BG
 
 | | A POTENTIAL ALLOSTERIC SUBSITE GENERATED BY DOMAIN SWAPPING IN BOVINE SEMINAL RIBONUCLEASE | | 分子名称: | PROTEIN (BOVINE SEMINAL RIBONUCLEASE), SULFATE ION, URIDYLYL-2'-5'-PHOSPHO-GUANOSINE | | 著者 | Vitagliano, L, Adinolfi, S, Sica, F, Merlino, A, Zagari, A, Mazzarella, L. | | 登録日 | 1999-03-11 | | 公開日 | 1999-11-05 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A potential allosteric subsite generated by domain swapping in bovine seminal ribonuclease.
J.Mol.Biol., 293, 1999
|
|
3WRQ
 
 | |
