4U5D
 
 | | Crystal structure of GluA2, con-ikot-ikot snail toxin, partial agonist KA and postitive modulator (R,R)-2b complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Con-ikot-ikot, ... | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-25 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.5757 Å) | | Cite: | X-ray structures of AMPA receptor-cone snail toxin complexes illuminate activation mechanism.
Science, 345, 2014
|
|
3VPM
 
 | | Crystal structure of human ribonucleotide reductase subunit M2 (hRRM2) mutant | | Descriptor: | FE (III) ION, MAGNESIUM ION, Ribonucleoside-diphosphate reductase subunit M2 | | Authors: | Chen, X, Xu, Z, Liu, H, Zhang, L, Chen, B, Zhu, L, Yang, C, Zhu, W, Shao, J. | | Deposit date: | 2012-03-05 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Essential role of E106 in the proton-coupled electron transfer in human ribonucleotide reductase M2 subunit
To be Published
|
|
4U5G
 
 | | Crystal structure of con-ikot-ikot toxin | | Descriptor: | Con-ikot-ikot, ZINC ION | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-25 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1997 Å) | | Cite: | X-ray structures of AMPA receptor-cone snail toxin complexes illuminate activation mechanism.
Science, 345, 2014
|
|
4U1Z
 
 | | GluA2flip sLBD complexed with kainate and (R,R)-2b crystal form D | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 2,Glutamate receptor 2, N,N'-[biphenyl-4,4'-diyldi(2R)propane-2,1-diyl]dipropane-2-sulfonamide | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-16 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9401 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
4U21
 
 | | GluA2flip sLBD complexed with FW and (R,R)-2b crystal form E | | Descriptor: | 2-AMINO-3-(5-FLUORO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, Glutamate receptor 2,Glutamate receptor 2, N,N'-[biphenyl-4,4'-diyldi(2R)propane-2,1-diyl]dipropane-2-sulfonamide | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-16 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.3908 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
4U5H
 
 | | crystal structure of con-ikot-ikot toxin | | Descriptor: | Con-ikot-ikot | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-25 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | X-ray structures of AMPA receptor-cone snail toxin complexes illuminate activation mechanism.
Science, 345, 2014
|
|
4U22
 
 | | GluA2flip sLBD complexed with FW and (R,R)-2b crystal form D | | Descriptor: | 2-AMINO-3-(5-FLUORO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, Glutamate receptor 2, N,N'-[biphenyl-4,4'-diyldi(2R)propane-2,1-diyl]dipropane-2-sulfonamide | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-16 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4409 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
4U23
 
 | | GluA2flip sLBD complexed with FW and (R,R)-2b crystal form F | | Descriptor: | 2-AMINO-3-(5-FLUORO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, Glutamate receptor 2,Glutamate receptor 2, N,N'-[biphenyl-4,4'-diyldi(2R)propane-2,1-diyl]dipropane-2-sulfonamide | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-16 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6734 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
7AF0
 
 | | Structure of SARS-CoV-2 Main Protease bound to Ipidacrine. | | Descriptor: | 2,3,5,6,7,8-hexahydro-1~{H}-cyclopenta[b]quinolin-9-amine, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-18 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7APH
 
 | | Structure of SARS-CoV-2 Main Protease bound to Tofogliflozin. | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-10-16 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
4UFU
 
 | | Crystal structure of human tankyrase 2 in complex with TA-12 | | Descriptor: | 8-methyl-2-[4-(trifluoromethyl)phenyl]-3H-quinazolin-4-one, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Lehtio, L. | | Deposit date: | 2015-03-19 | | Release date: | 2016-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Activity Relationships of 2-Arylquinazolin-4-Ones as Highly Selective and Potent Inhibitors of the Tankyrases.
Eur.J.Med.Chem., 118, 2016
|
|
4TVA
 
 | | Universal Pathway for Post-Transfer Editing Reactions: Insight from Crystal structure of TthPheRS with Puromycine | | Descriptor: | PHENYLALANINE, PUROMYCIN, Phenylalanine--tRNA ligase alpha subunit, ... | | Authors: | Safro, M, Klipcan, L, Tworowski, D, Peretz, M. | | Deposit date: | 2014-06-26 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | Universal pathway for posttransfer editing reactions: Insights from the crystal structure of TtPheRS with puromycin.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4TXN
 
 | | Crystal structure of uridine phosphorylase from Schistosoma mansoni in complex with 5-fluorouracil | | Descriptor: | 5-FLUOROURACIL, SULFATE ION, Uridine phosphorylase | | Authors: | Marinho, A, Torini, J, Romanello, L, Cassago, A, DeMarco, R, Brandao-Neto, J, Pereira, H.M. | | Deposit date: | 2014-07-03 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analysis of two Schistosoma mansoni uridine phosphorylases isoforms suggests the emergence of a protein with a non-canonical function.
Biochimie, 125, 2016
|
|
4U1Y
 
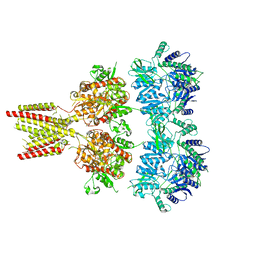 | | Full length GluA2-FW-(R,R)-2b complex | | Descriptor: | 2-AMINO-3-(5-FLUORO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2, ... | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-16 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.8999 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
4TXH
 
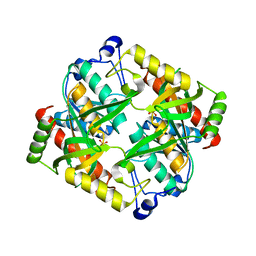 | | Crystal structure of uridine phosphorylase from Schistosoma mansoni in APO form | | Descriptor: | SULFATE ION, Uridine phosphorylase | | Authors: | Torini, J, Romanello, L, Marinho, A, Brandao-Neto, J, Cassago, A, DeMarco, R, Pereira, H.M. | | Deposit date: | 2014-07-03 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Analysis of two Schistosoma mansoni uridine phosphorylases isoforms suggests the emergence of a protein with a non-canonical function.
Biochimie, 125, 2016
|
|
7AMJ
 
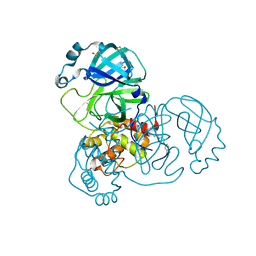 | | Structure of SARS-CoV-2 Main Protease bound to PD 168568. | | Descriptor: | (3~{S})-3-[2-[4-(3,4-dimethylphenyl)piperazin-1-yl]ethyl]-2,3-dihydroisoindol-1-one, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Hinrichs, W, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Boger, J, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-10-09 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
4TUT
 
 | | Structure of a Prion peptide | | Descriptor: | Prion peptide: GLY-GLY-TYR-MET-LEU-GLY | | Authors: | Yu, L, Lee, S.-J, Yee, V. | | Deposit date: | 2014-06-24 | | Release date: | 2015-05-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Crystal Structures of Polymorphic Prion Protein beta 1 Peptides Reveal Variable Steric Zipper Conformations.
Biochemistry, 54, 2015
|
|
7AK4
 
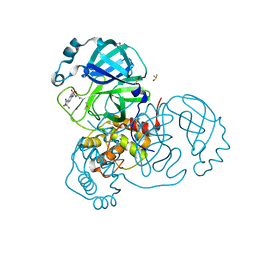 | | Structure of SARS-CoV-2 Main Protease bound to Tretazicar. | | Descriptor: | 3C-like proteinase, 5-(AZIRIDIN-1-YL)-2,4-DINITROBENZAMIDE, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AX6
 
 | | Structure of SARS-CoV-2 Main Protease bound to Glutathione isopropyl ester | | Descriptor: | (2~{S})-2-azanyl-5-oxidanylidene-5-[[(2~{S})-1-oxidanylidene-1-[(2-oxidanylidene-2-propan-2-yloxy-ethyl)amino]-3-sulfanyl-propan-2-yl]amino]pentanoic acid, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-11-09 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
4TXJ
 
 | | Crystal structure of uridine phosphorylase from Schistosoma mansoni in complex with thymidine | | Descriptor: | SULFATE ION, THYMIDINE, Uridine phosphorylase | | Authors: | Torini, J, Marinho, A, Romanello, L, Cassago, A, DeMarco, R, Brandao-Neto, J, Pereira, H.M. | | Deposit date: | 2014-07-03 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.662 Å) | | Cite: | Analysis of two Schistosoma mansoni uridine phosphorylases isoforms suggests the emergence of a protein with a non-canonical function.
Biochimie, 125, 2016
|
|
7AKU
 
 | | Structure of SARS-CoV-2 Main Protease bound to Calpeptin. | | Descriptor: | 3C-like proteinase, CHLORIDE ION, Calpeptin | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-10-02 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
4U01
 
 | | HCV NS3/4A serine protease in complex with 6570 | | Descriptor: | (2S,3aS,10Z,11aS,12aR)-2-({8-fluoro-7-methoxy-2-[4-(propan-2-yl)-1,3-thiazol-2-yl]quinolin-4-yl}oxy)-5-methyl-N-[(1-methylcyclopropyl)sulfonyl]-4,14-dioxo-1,2,3,3a,4,5,6,7,8,9,11a,12,13,14-tetradecahydro-12aH-cyclopropa[m]pyrrolo[1,2-c][1,3,6]triazacyclotetradecine-12a-carboxamide, CHLORIDE ION, NS4A protein, ... | | Authors: | Parsy, C.C, Alexandre, F.-R, Brandt, G, Caillet, C, Chaves, D, Derock, M, Gloux, D, Griffon, Y, Lallos, L.B, Leroy, F, Liuzzi, M, Loi, A.-G, Mayes, B, Moulat, L, Moussa, A, Chiara, M, Roques, V, Rosinovsky, E, Seifer, M, Stewart, A, Wang, J, Standring, D, Surleraux, D. | | Deposit date: | 2014-07-11 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and structural diversity of the hepatitis C virus NS3/4A serine protease inhibitor series leading to clinical candidate IDX320.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
7AWR
 
 | | Structure of SARS-CoV-2 Main Protease bound to Tegafur | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, TEGAFUR | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-11-09 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
4U1O
 
 | | GluA2flip sLBD complexed with kainate and (R,R)-2b crystal form C | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 2, N,N'-[biphenyl-4,4'-diyldi(2R)propane-2,1-diyl]dipropane-2-sulfonamide | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-15 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8501 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
7ARF
 
 | | Structure of SARS-CoV-2 Main Protease bound to thioglucose. | | Descriptor: | (2~{S},3~{R},4~{R},5~{S},6~{S})-2-(hydroxymethyl)-6-sulfanyl-oxane-3,4,5-triol, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-10-24 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
