6F9O
 
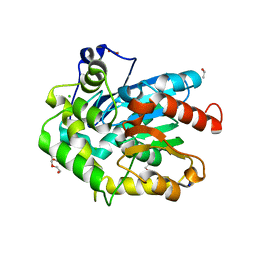 | | Crystal structure of cold-adapted haloalkane dehalogenase DpcA from Psychrobacter cryohalolentis K5 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Haloalkane dehalogenase, ... | | Authors: | Tratsiak, K, Prudnikova, T, Drienovska, I, Damborsky, J, Brynda, J, Pachl, P, Kuty, M, Chaloupkova, R, Kuta Smatanova, I, Rezacova, P. | | Deposit date: | 2017-12-15 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Crystal structure of the cold-adapted haloalkane dehalogenase DpcA from Psychrobacter cryohalolentis K5.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6IAH
 
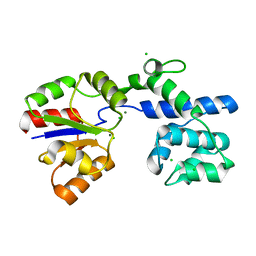 | | Phosphatase Tt82 from Thermococcus thioreducens | | Descriptor: | CHLORIDE ION, Hydrolase, MAGNESIUM ION | | Authors: | Havlickova, P, Brinsa, V, Brynda, J, Pachl, P, Prudnikova, T, Mesters, J.R, Kascakova, B, Kuty, M, Pusey, M.L, Ng, J.D, Rezacova, P, Smatanova, I.K. | | Deposit date: | 2018-11-26 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A novel structurally characterized haloacid dehalogenase superfamily phosphatase from Thermococcus thioreducens with diverse substrate specificity.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
3ZHO
 
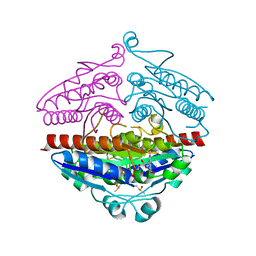 | | X-ray structure of E.coli Wrba in complex with FMN at 1.2 A resolution | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVOPROTEIN WRBA | | Authors: | Kishko, I, Lapkouski, M, Brynda, J, Kuty, M, Carey, J, Smatanova, I.K, Ettrich, R. | | Deposit date: | 2012-12-23 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | 1.2 A Resolution Crystal Structure of Escherichia Coli Wrba Holoprotein
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4K2A
 
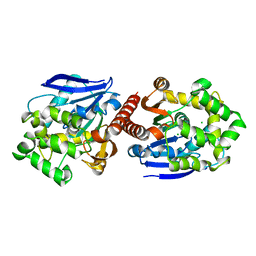 | | Crystal structure of haloalkane dehalogenase DbeA from Bradyrhizobium elkani USDA94 | | Descriptor: | ACETATE ION, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Prudnikova, T, Chaloupkova, R, Rezacova, P, Mozga, T, Koudelakova, T, Sato, Y, Kuty, M, Nagata, Y, Damborsky, J, Kuta Smatanova, I, Structure 2 Function Project (S2F) | | Deposit date: | 2013-04-08 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional analysis of a novel haloalkane dehalogenase with two halide-binding sites.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3FWH
 
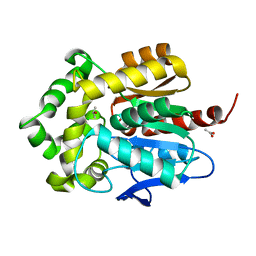 | | Structure of haloalkane dehalogenase mutant Dha15 (I135F/C176Y) from Rhodococcus rhodochrous | | Descriptor: | ACETATE ION, CHLORIDE ION, Haloalkane dehalogenase, ... | | Authors: | Gavira, J.A, Stsiapanava, A, Kuty, M, Dohnalek, J, Lapkouski, M, Kuta Smatanova, I. | | Deposit date: | 2009-01-18 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Atomic resolution studies of haloalkane dehalogenases DhaA04, DhaA14 and DhaA15 with engineered access tunnels.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3G9X
 
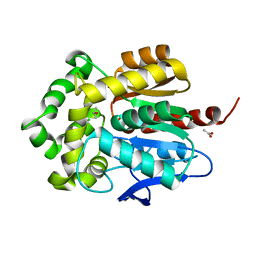 | | Structure of haloalkane dehalogenase DhaA14 mutant I135F from Rhodococcus rhodochrous | | Descriptor: | ACETATE ION, CHLORIDE ION, Haloalkane dehalogenase, ... | | Authors: | Gavira, J.A, Stsiapanava, A, Kuty, M, Lapkouski, M, Dohnalek, J, Kuta Smatanova, I. | | Deposit date: | 2009-02-15 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Atomic resolution studies of haloalkane dehalogenases DhaA04, DhaA14 and DhaA15 with engineered access tunnels.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3FBW
 
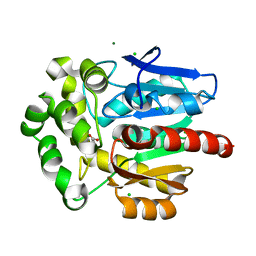 | | Structure of Rhodococcus rhodochrous haloalkane dehalogenase DhaA mutant C176Y | | Descriptor: | BENZOIC ACID, CHLORIDE ION, Haloalkane dehalogenase, ... | | Authors: | Dohnalek, J, Stsiapanava, A, Gavira, J.A, Kuta Smatanova, I, Kuty, M. | | Deposit date: | 2008-11-20 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Atomic resolution studies of haloalkane dehalogenases DhaA04, DhaA14 and DhaA15 with engineered access tunnels.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1PDX
 
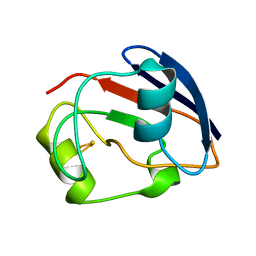 | | PUTIDAREDOXIN | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, PROTEIN (PUTIDAREDOXIN) | | Authors: | Pochapsky, T.C, Jain, N.U, Kuti, M, Lyons, T.A, Heymont, J. | | Deposit date: | 1999-02-15 | | Release date: | 1999-05-12 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A refined model for the solution structure of oxidized putidaredoxin.
Biochemistry, 38, 1999
|
|
1GPX
 
 | | C85S GAPDX, NMR, 20 STRUCTURES | | Descriptor: | GALLIUM (III) ION, PUTIDAREDOXIN | | Authors: | Pochapsky, T.C, Kuti, M, Kazanis, S. | | Deposit date: | 1998-06-10 | | Release date: | 1999-01-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a gallium-substituted putidaredoxin mutant: GaPdx C85S.
J.Biomol.NMR, 12, 1998
|
|
1XR0
 
 | | Structural Basis of SNT PTB Domain Interactions with Distinct Neurotrophic Receptors | | Descriptor: | Basic fibroblast growth factor receptor 1, FGFR signalling adaptor SNT-1 | | Authors: | Dhalluin, C, Yan, K.S, Plotnikova, O, Lee, K.W, Zeng, L, Kuti, M, Mujtaba, S, Goldfarb, M.P, Zhou, M.-M. | | Deposit date: | 2004-10-13 | | Release date: | 2004-11-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of SNT PTB Domain Interactions with Distinct Neurotrophic Receptors
Mol.Cell, 6, 2000
|
|
