3WS5
 
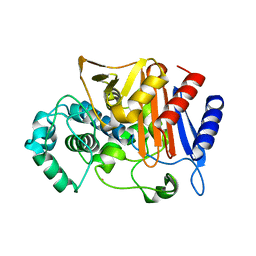 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (Condition-2B) | | Descriptor: | Beta-lactamase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-28 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WRZ
 
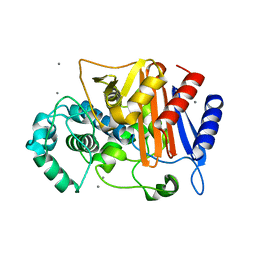 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (without soaking) | | Descriptor: | Beta-lactamase, CALCIUM ION, CHLORIDE ION | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WS0
 
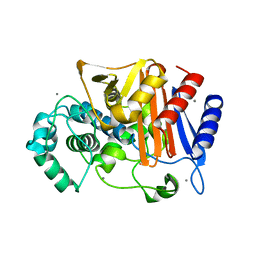 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (Condition-1A) | | Descriptor: | Beta-lactamase, CALCIUM ION, CESIUM ION, ... | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3VGH
 
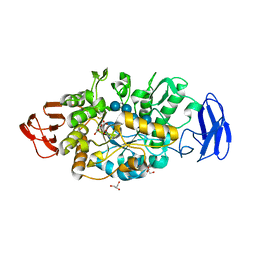 | | Crystal structure of glycosyltrehalose trehalohydrolase (E283Q) complexed with maltotriosyltrehalose | | Descriptor: | CITRATE ANION, GLYCEROL, Malto-oligosyltrehalose trehalohydrolase, ... | | Authors: | Okazaki, N, Tamada, T, Feese, M.D, Kato, M, Miura, Y, Komeda, T, Kobayashi, K, Kondo, K, Kuroki, R. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Substrate recognition mechanism of a glycosyltrehalose trehalohydrolase from Sulfolobus solfataricus KM1.
Protein Sci., 21, 2012
|
|
3VXF
 
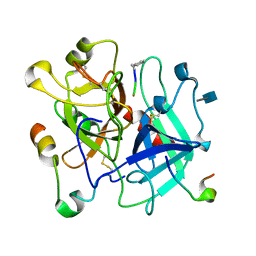 | | X/N Joint refinement of Human alpha-thrombin-Bivalirudin complex PD5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BIVALIRUDIN, Thrombin heavy chain, ... | | Authors: | Yamada, T, Kurihara, K, Masumi, K, Tamada, T, Tomoyori, K, Ohnishi, Y, Tanaka, I, Kuroki, R, Niimura, N. | | Deposit date: | 2012-09-12 | | Release date: | 2013-09-04 | | Last modified: | 2020-07-29 | | Method: | NEUTRON DIFFRACTION (1.602 Å), X-RAY DIFFRACTION | | Cite: | Neutron and X-ray crystallographic analysis of the human alpha-thrombin-bivalirudin complex at pD 5.0: protonation states and hydration structure of the enzyme-product complex
Biochim.Biophys.Acta, 1834, 2013
|
|
3VGS
 
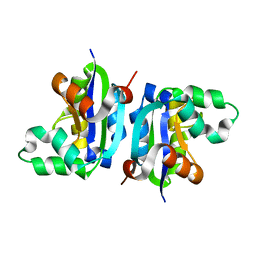 | | Wild-type nucleoside diphosphate kinase derived from Halomonas sp. 593 | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Okazaki, N, Yonezawa, Y, Arai, S, Matsumoto, F, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2011-08-20 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A structural mechanism for dimeric to tetrameric oligomer conversion in Halomonas sp. nucleoside diphosphate kinase
Protein Sci., 21, 2012
|
|
3VGG
 
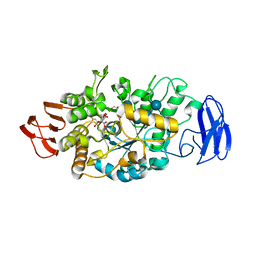 | | Crystal structure of glycosyltrehalose trehalohydrolase (E283Q) complexed with maltoheptaose | | Descriptor: | CITRATE ANION, GLYCEROL, Malto-oligosyltrehalose trehalohydrolase, ... | | Authors: | Okazaki, N, Tamada, T, Feese, M.D, Kato, M, Miura, Y, Komeda, T, Kobayashi, K, Kondo, K, Kuroki, R. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Substrate recognition mechanism of a glycosyltrehalose trehalohydrolase from Sulfolobus solfataricus KM1.
Protein Sci., 21, 2012
|
|
3VGT
 
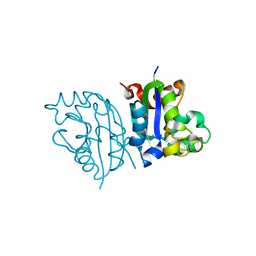 | | Wild-type nucleoside diphosphate kinase derived from Halomonas sp. 593 | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Okazaki, N, Yonezawa, Y, Arai, S, Matsumoto, F, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2011-08-21 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A structural mechanism for dimeric to tetrameric oligomer conversion in Halomonas sp. nucleoside diphosphate kinase
Protein Sci., 21, 2012
|
|
3VXE
 
 | | Human alpha-thrombin-Bivalirudin complex at PD5.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BIVALIRUDIN, Thrombin heavy chain, ... | | Authors: | Yamada, T, Kurihara, K, Masumi, K, Tamada, T, Tomoyori, K, Ohnishi, Y, Tanaka, I, Kuroki, R, Niimura, N. | | Deposit date: | 2012-09-12 | | Release date: | 2013-09-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Neutron and X-ray crystallographic analysis of the human alpha-thrombin-bivalirudin complex at pD 5.0: protonation states and hydration structure of the enzyme-product complex
Biochim.Biophys.Acta, 1834, 2013
|
|
3W2E
 
 | | Crystal structure of oxidation intermediate (20 min) of NADH-cytochrome b5 reductase from pig liver | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH-cytochrome b5 reductase 3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamada, M, Tamada, T, Matsumoto, F, Shoyama, Y, Kimura, S, Kuroki, R, Miki, K. | | Deposit date: | 2012-11-28 | | Release date: | 2013-07-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Elucidations of the catalytic cycle of NADH-cytochrome b5 reductase by X-ray crystallography: new insights into regulation of efficient electron transfer
J.Mol.Biol., 425, 2013
|
|
3W2G
 
 | | Crystal structure of fully reduced form of NADH-cytochrome b5 reductase from pig liver | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH-cytochrome b5 reductase 3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamada, M, Tamada, T, Matsumoto, F, Shoyama, Y, Kimura, S, Kuroki, R, Miki, K. | | Deposit date: | 2012-11-28 | | Release date: | 2013-07-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Elucidations of the catalytic cycle of NADH-cytochrome b5 reductase by X-ray crystallography: new insights into regulation of efficient electron transfer
J.Mol.Biol., 425, 2013
|
|
3W2F
 
 | | Crystal structure of oxidation intermediate (10 min) of NADH-cytochrome b5 reductase from pig liver | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH-cytochrome b5 reductase 3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamada, M, Tamada, T, Matsumoto, F, Shoyama, Y, Kimura, S, Kuroki, R, Miki, K. | | Deposit date: | 2012-11-28 | | Release date: | 2013-07-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Elucidations of the catalytic cycle of NADH-cytochrome b5 reductase by X-ray crystallography: new insights into regulation of efficient electron transfer
J.Mol.Biol., 425, 2013
|
|
3WBH
 
 | | Structural characteristics of alkaline phosphatase from a moderately halophilic bacteria Halomonas sp.593 | | Descriptor: | Alkaline phosphatase, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Arai, S, Yonezawa, Y, Ishibashi, M, Matsumoto, F, Tamada, T, Tokunaga, H, Tokunaga, M, Kuroki, R. | | Deposit date: | 2013-05-17 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characteristics of alkaline phosphatase from the moderately halophilic bacterium Halomonas sp. 593.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3W2H
 
 | | Crystal structure of oxidation intermediate (1min) of NADH-cytochrome b5 reductase from pig liver | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH-cytochrome b5 reductase 3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamada, M, Tamada, T, Matsumoto, F, Shoyama, Y, Kimura, S, Kuroki, R, Miki, K. | | Deposit date: | 2012-11-28 | | Release date: | 2013-07-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Elucidations of the catalytic cycle of NADH-cytochrome b5 reductase by X-ray crystallography: new insights into regulation of efficient electron transfer
J.Mol.Biol., 425, 2013
|
|
3VGB
 
 | | Crystal structure of glycosyltrehalose trehalohydrolase (GTHase) from Sulfolobus solfataricus KM1 | | Descriptor: | CITRATE ANION, GLYCEROL, Malto-oligosyltrehalose trehalohydrolase | | Authors: | Okazaki, N, Tamada, T, Feese, M.D, Kato, M, Miura, Y, Komeda, T, Kobayashi, K, Kondo, K, Kuroki, R. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Substrate recognition mechanism of a glycosyltrehalose trehalohydrolase from Sulfolobus solfataricus KM1.
Protein Sci., 21, 2012
|
|
3VGU
 
 | | E134A mutant nucleoside diphosphate kinase derived from Halomonas sp. 593 | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Okazaki, N, Yonezawa, Y, Arai, S, Matsumoto, F, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2011-08-21 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A structural mechanism for dimeric to tetrameric oligomer conversion in Halomonas sp. nucleoside diphosphate kinase
Protein Sci., 21, 2012
|
|
3VGE
 
 | | Crystal structure of glycosyltrehalose trehalohydrolase (D252S) | | Descriptor: | CITRATE ANION, GLYCEROL, Malto-oligosyltrehalose trehalohydrolase | | Authors: | Okazaki, N, Tamada, T, Feese, M.D, Kato, M, Miura, Y, Komeda, T, Kobayashi, K, Kondo, K, Kuroki, R. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Substrate recognition mechanism of a glycosyltrehalose trehalohydrolase from Sulfolobus solfataricus KM1.
Protein Sci., 21, 2012
|
|
3VGD
 
 | | Ctystal structure of glycosyltrehalose trehalohydrolase (D252E) | | Descriptor: | CITRATE ANION, GLYCEROL, Malto-oligosyltrehalose trehalohydrolase | | Authors: | Okazaki, N, Tamada, T, Feese, M.D, Kato, M, Miura, Y, Komeda, T, Kobayashi, K, Kondo, K, Kuroki, R. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Substrate recognition mechanism of a glycosyltrehalose trehalohydrolase from Sulfolobus solfataricus KM1.
Protein Sci., 21, 2012
|
|
3VGF
 
 | | Crystal structure of glycosyltrehalose trehalohydrolase (D252S) complexed with maltotriosyltrehalose | | Descriptor: | CITRATE ANION, GLYCEROL, Malto-oligosyltrehalose trehalohydrolase, ... | | Authors: | Okazaki, N, Tamada, T, Feese, M.D, Kato, M, Miura, Y, Komeda, T, Kobayashi, K, Kondo, K, Kuroki, R. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrate recognition mechanism of a glycosyltrehalose trehalohydrolase from Sulfolobus solfataricus KM1.
Protein Sci., 21, 2012
|
|
3VGV
 
 | | E134A mutant nucleoside diphosphate kinase derived from Halomonas sp. 593 | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Okazaki, N, Yonezawa, Y, Arai, S, Matsumoto, F, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2011-08-21 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A structural mechanism for dimeric to tetrameric oligomer conversion in Halomonas sp. nucleoside diphosphate kinase
Protein Sci., 21, 2012
|
|
3WA2
 
 | | High resolution crystal structure of copper amine oxidase from arthrobacter globiformis | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Murakawa, T, Hayashi, H, Sunami, T, Kurihara, K, Tamada, T, Kuroki, R, Suzuki, M, Tanizawa, K, Okajima, T. | | Deposit date: | 2013-04-22 | | Release date: | 2013-09-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | High-resolution crystal structure of copper amine oxidase from Arthrobacter globiformis: assignment of bound diatomic molecules as O2
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3W2I
 
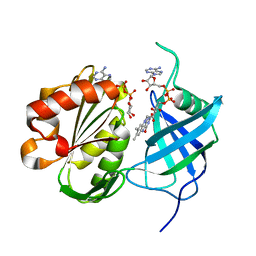 | | Crystal structure of re-oxidized form (60 min) of NADH-cytochrome b5 reductase from pig liver | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH-cytochrome b5 reductase 3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamada, M, Tamada, T, Matsumoto, F, Shoyama, Y, Kimura, S, Kuroki, R, Miki, K. | | Deposit date: | 2012-11-28 | | Release date: | 2013-07-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Elucidations of the catalytic cycle of NADH-cytochrome b5 reductase by X-ray crystallography: new insights into regulation of efficient electron transfer
J.Mol.Biol., 425, 2013
|
|
3WA3
 
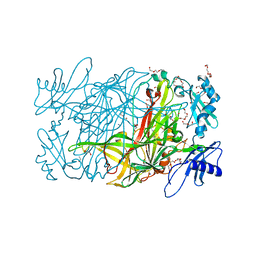 | | Crystal structure of copper amine oxidase from arthrobacter globiformis in N2 condition | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Murakawa, T, Hayashi, H, Sunami, T, Kurihara, K, Tamada, T, Kuroki, R, Suzuki, M, Tanizawa, K, Okajima, T. | | Deposit date: | 2013-04-22 | | Release date: | 2013-09-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | High-resolution crystal structure of copper amine oxidase from Arthrobacter globiformis: assignment of bound diatomic molecules as O2
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3WC4
 
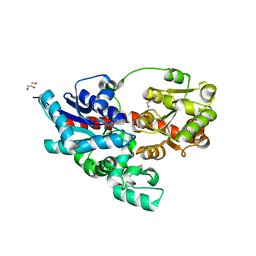 | | Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea | | Descriptor: | ACETATE ION, GLYCEROL, UDP-glucose:anthocyanidin 3-O-glucosyltransferase | | Authors: | Hiromoto, T, Honjo, E, Tamada, T, Kuroki, R. | | Deposit date: | 2013-05-24 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of UDP-glucose:anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea
J.SYNCHROTRON RADIAT., 20, 2013
|
|
