2QQ3
 
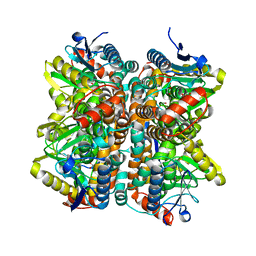 | | Crystal Structure Of Enoyl-CoA Hydrates Subunit I (gk_2039) Other Form From Geobacillus Kaustophilus HTA426 | | Descriptor: | 1,2-ETHANEDIOL, Enoyl-CoA hydratase subunit I | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Sekar, K, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-07-26 | | Release date: | 2008-07-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure Of Enoyl-CoA Hydrates Subunit I (gk_2039) Other Form From Geobacillus Kaustophilus HTA426
To be Published
|
|
3CM0
 
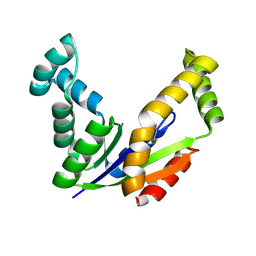 | | Crystal structure of adenylate kinase from Thermus thermophilus HB8 | | Descriptor: | Adenylate kinase | | Authors: | Nakagawa, N, Kondo, N, Masui, R, Matsuda, Z, Iwamoto, I, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-03-20 | | Release date: | 2008-09-23 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of adenylate kinase from Thermus thermophilus HB8.
To be Published
|
|
3VPR
 
 | | Crystal Structure of a TetR Family Transcriptional Regulator PfmR from Thermus thermophilus HB8 | | Descriptor: | Transcriptional regulator, TetR family | | Authors: | Agari, Y, Sakamoto, K, Kuramitsu, S, Shinkai, A. | | Deposit date: | 2012-03-12 | | Release date: | 2012-07-11 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Transcriptional repression mediated by a TetR family protein, PfmR, from Thermus thermophilus HB8
J.Bacteriol., 2012
|
|
3CPQ
 
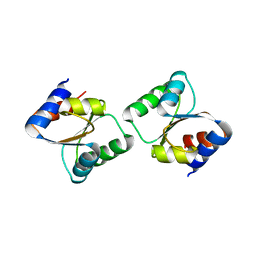 | | Crystal Structure of L30e a ribosomal protein from Methanocaldococcus jannaschii DSM2661 (MJ1044) | | Descriptor: | 50S ribosomal protein L30e | | Authors: | Jeyakanthan, J, Sarani, R, Mridula, P, Sekar, K, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-04-01 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of L30e a ribosomal protein from Methanocaldococcus jannaschii DSM2661 (MJ1044)
To be Published
|
|
1UKW
 
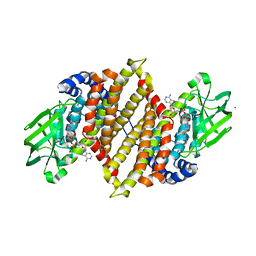 | | Crystal structure of medium-chain acyl-CoA dehydrogenase from Thermus thermophilus HB8 | | Descriptor: | COBALT (II) ION, FLAVIN-ADENINE DINUCLEOTIDE, acyl-CoA dehydrogenase | | Authors: | Hamada, K, Ago, H, Kuramitsu, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-02 | | Release date: | 2004-11-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Thermus thermophilus medium-chain acyl-CoA dehydrogenase
To be published
|
|
3W8N
 
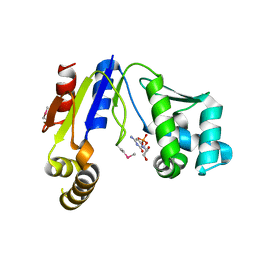 | |
1VCD
 
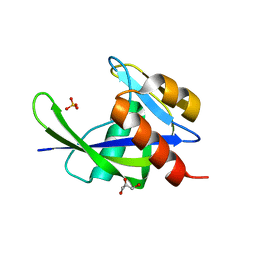 | | Crystal Structure of a T.thermophilus HB8 Ap6A hydrolase Ndx1 | | Descriptor: | GLYCEROL, Ndx1, SULFATE ION | | Authors: | Iwai, T, Nakagawa, N, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-05 | | Release date: | 2005-04-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Nudix Protein Ndx1 from Thermus thermophilus HB8 in binary complex with diadenosine hexaphosphate
To be Published
|
|
1C9C
 
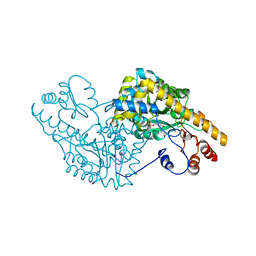 | | ASPARTATE AMINOTRANSFERASE COMPLEXED WITH C3-PYRIDOXAL-5'-PHOSPHATE | | Descriptor: | ALANYL-PYRIDOXAL-5'-PHOSPHATE, ASPARTATE AMINOTRANSFERASE | | Authors: | Ishijima, J, Nakai, T, Kawaguchi, S, Hirotsu, K, Kuramitsu, S. | | Deposit date: | 1999-08-02 | | Release date: | 2000-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Free energy requirement for domain movement of an enzyme
J.Biol.Chem., 275, 2000
|
|
1CQ8
 
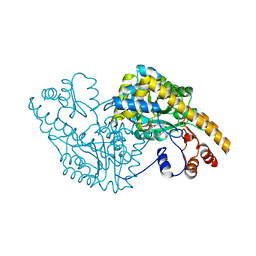 | | ASPARTATE AMINOTRANSFERASE (E.C. 2.6.1.1) COMPLEXED WITH C6-PYRIDOXAL-5P-PHOSPHATE | | Descriptor: | 2-[O-PHOSPHONOPYRIDOXYL]-AMINO-HEXANOIC ACID, ASPARTATE AMINOTRANSFERASE (2.6.1.1) | | Authors: | Ishijima, J, Nakai, T, Kawaguchi, S, Hirotsu, K, Kuramitsu, S. | | Deposit date: | 1999-08-06 | | Release date: | 2000-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Free energy requirement for domain movement of an enzyme
J.Biol.Chem., 275, 2000
|
|
1EE8
 
 | | CRYSTAL STRUCTURE OF MUTM (FPG) PROTEIN FROM THERMUS THERMOPHILUS HB8 | | Descriptor: | MUTM (FPG) PROTEIN, ZINC ION | | Authors: | Sugahara, M, Mikawa, T, Kumasaka, T, Yamamoto, M, Kato, R, Fukuyama, K, Inoue, Y, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2000-01-31 | | Release date: | 2001-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a repair enzyme of oxidatively damaged DNA, MutM (Fpg), from an extreme thermophile, Thermus thermophilus HB8.
EMBO J., 19, 2000
|
|
1D2M
 
 | | UVRB PROTEIN OF THERMUS THERMOPHILUS HB8; A NUCLEOTIDE EXCISION REPAIR ENZYME | | Descriptor: | EXCINUCLEASE ABC SUBUNIT B, SULFATE ION, octyl beta-D-glucopyranoside | | Authors: | Nakagawa, N, Sugahara, M, Masui, R, Kato, R, Fukuyama, K, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-09-25 | | Release date: | 2000-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Thermus thermophilus HB8 UvrB protein, a key enzyme of nucleotide excision repair.
J.Biochem.(Tokyo), 126, 1999
|
|
2QQ1
 
 | | Crystal Structure Of Molybdenum Cofactor Biosynthesis (aq_061) Other Form From Aquifex Aeolicus Vf5 | | Descriptor: | Molybdenum cofactor biosynthesis MOG | | Authors: | Jeyakanthan, J, Mahesh, S, Kanaujia, S.P, Ramakumar, S, Sekar, K, Agari, Y, Ebihara, A, Kuramitsu, S, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-07-26 | | Release date: | 2008-07-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure Of Molybdenum Cofactor Biosynthesis (aq_061) Other Form From Aquifex Aeolicus Vf5
To be Published
|
|
1NZ8
 
 | | Solution Structure of the N-utilization substance G (NusG) N-terminal (NGN) domain from Thermus thermophilus | | Descriptor: | TRANSCRIPTION ANTITERMINATION PROTEIN NUSG | | Authors: | Reay, P, Yamasaki, K, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-17 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and sequence comparisons arising from the solution structure of the transcription elongation factor NusG from Thermus thermophilus
Proteins, 56, 2004
|
|
1NZ9
 
 | | Solution Structure of the N-utilization substance G (NusG) C-terminal (NGC) domain from Thermus thermophilus | | Descriptor: | TRANSCRIPTION ANTITERMINATION PROTEIN NUSG | | Authors: | Reay, P, Yamasaki, K, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-17 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and sequence comparisons arising from the solution structure of the transcription elongation factor NusG from Thermus thermophilus
Proteins, 56, 2004
|
|
3CQ1
 
 | | Structure of the DTDP-4-Keto-L-Rhamnose Reductase related protein (TT1362) from Thermus Thermophilus HB8 | | Descriptor: | Putative uncharacterized protein TTHB138 | | Authors: | Jeyakanthan, J, Satoh, S, Kitamura, Y, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-04-02 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the DTDP-4-Keto-L-Rhamnose Reductase related protein (TT1362) from Thermus Thermophilus HB8
To be Published
|
|
1UMD
 
 | | branched-chain 2-oxo acid dehydrogenase (E1) from Thermus thermophilus HB8 with 4-methyl-2-oxopentanoate as an intermediate | | Descriptor: | 2-OXO-4-METHYLPENTANOIC ACID, 2-oxo acid dehydrogenase alpha subunit, 2-oxo acid dehydrogenase beta subunit, ... | | Authors: | Nakai, T, Nakagawa, N, Maoka, N, Masui, R, Kuramitsu, S, Kamiya, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-25 | | Release date: | 2004-03-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ligand-induced Conformational Changes and a Reaction Intermediate in Branched-chain 2-Oxo Acid Dehydrogenase (E1) from Thermus thermophilus HB8, as Revealed by X-ray Crystallography
J.Mol.Biol., 337, 2004
|
|
2WQK
 
 | | Crystal Structure of Sure Protein from Aquifex aeolicus | | Descriptor: | 5'-NUCLEOTIDASE SURE, SODIUM ION, SULFATE ION | | Authors: | Antonyuk, S.V, Ellis, M.J, Strange, R.W, Hasnain, S.S, Bessho, Y, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-08-23 | | Release date: | 2009-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Sure Protein from Aquifex Aeolicus Vf5 at 1.5 A Resolution.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
1UMB
 
 | | branched-chain 2-oxo acid dehydrogenase (E1) from Thermus thermophilus HB8 in holo-form | | Descriptor: | 2-oxo acid dehydrogenase alpha subunit, 2-oxo acid dehydrogenase beta subunit, MAGNESIUM ION, ... | | Authors: | Nakai, T, Nakagawa, N, Maoka, N, Masui, R, Kuramitsu, S, Kamiya, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-25 | | Release date: | 2004-03-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ligand-induced Conformational Changes and a Reaction Intermediate in Branched-chain 2-Oxo Acid Dehydrogenase (E1) from Thermus thermophilus HB8, as Revealed by X-ray Crystallography
J.Mol.Biol., 337, 2004
|
|
1UM9
 
 | | branched-chain 2-oxo acid dehydrogenase (E1) from Thermus thermophilus HB8 in apo-form | | Descriptor: | 2-oxo acid dehydrogenase alpha subunit, 2-oxo acid dehydrogenase beta subunit, SULFATE ION | | Authors: | Nakai, T, Nakagawa, N, Maoka, N, Masui, R, Kuramitsu, S, Kamiya, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-25 | | Release date: | 2004-03-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ligand-induced Conformational Changes and a Reaction Intermediate in Branched-chain 2-Oxo Acid Dehydrogenase (E1) from Thermus thermophilus HB8, as Revealed by X-ray Crystallography
J.Mol.Biol., 337, 2004
|
|
1UMC
 
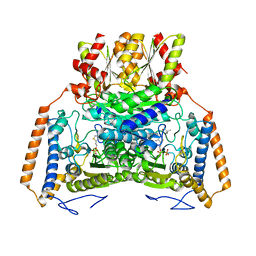 | | branched-chain 2-oxo acid dehydrogenase (E1) from Thermus thermophilus HB8 with 4-methylpentanoate | | Descriptor: | 2-oxo acid dehydrogenase alpha subunit, 2-oxo acid dehydrogenase beta subunit, 4-METHYL VALERIC ACID, ... | | Authors: | Nakai, T, Nakagawa, N, Maoka, N, Masui, R, Kuramitsu, S, Kamiya, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-25 | | Release date: | 2004-03-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ligand-induced Conformational Changes and a Reaction Intermediate in Branched-chain 2-Oxo Acid Dehydrogenase (E1) from Thermus thermophilus HB8, as Revealed by X-ray Crystallography
J.Mol.Biol., 337, 2004
|
|
1UFR
 
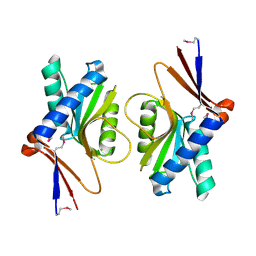 | | Crystal Structure of TT1027 from Thermus thermophilus HB8 | | Descriptor: | CHLORIDE ION, pyr mRNA-binding attenuation protein | | Authors: | Matsuura, T, Sakai, H, Terada, T, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-08 | | Release date: | 2003-12-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of TT1027 from Thermus thermophilus HB8
To be Published
|
|
1UF9
 
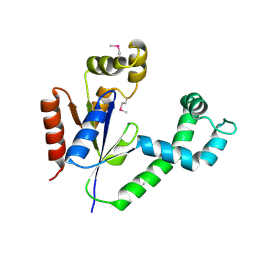 | | Crystal structure of TT1252 from Thermus thermophilus | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, PHOSPHATE ION, TT1252 protein | | Authors: | Seto, A, Murayama, K, Toyama, M, Ebihara, A, Nakagawa, N, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-28 | | Release date: | 2003-11-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | ATP-induced structural change of dephosphocoenzyme A kinase from Thermus thermophilus HB8
PROTEINS, 58, 2005
|
|
1UFV
 
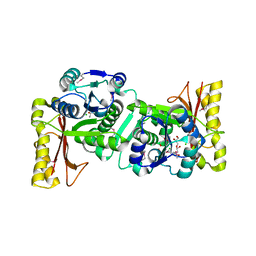 | | Crystal Structure Of Pantothenate Synthetase From Thermus Thermophilus HB8 | | Descriptor: | CHLORIDE ION, GLYCEROL, Pantoate-beta-alanine ligase | | Authors: | Bagautdinov, B, Kuramitsu, S, Yokoyama, S, Miyano, M, Tahirov, T.H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-10 | | Release date: | 2003-06-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure Of Pantothenate Synthetase From Thermus Thermophilus HB8
To be Published
|
|
1OI7
 
 | | The Crystal Structure of Succinyl-CoA synthetase alpha subunit from Thermus Thermophilus | | Descriptor: | SUCCINYL-COA SYNTHETASE ALPHA CHAIN | | Authors: | Takahashi, H, Tokunaga, Y, Kuroishi, C, Babayeva, N, Kuramitsu, S, Yokoyama, S, Miyano, M, Tahirov, T.H. | | Deposit date: | 2003-06-10 | | Release date: | 2003-07-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | The Crystal Structure of Succinyl-Coa Synthetase from Thermus Thermophilus
To be Published
|
|
1UFA
 
 | | Crystal structure of TT1467 from Thermus thermophilus HB8 | | Descriptor: | TT1467 protein | | Authors: | Idaka, M, Terada, T, Murayama, K, Yamaguchi, H, Nureki, O, Ishitani, R, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-28 | | Release date: | 2003-11-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of TT1467 from Thermus thermophilus HB8
To be published
|
|
