2YX2
 
 | | Crystal structure of cloned trimeric hyluranidase from streptococcus pyogenes at 2.8 A resolution | | Descriptor: | Hyaluronidase, phage associated | | Authors: | Mishra, P, Prem Kumar, R, Bhakuni, V, Singh, N, Sharma, S, Kaur, P, Perbandt, M, Betzel, C, Singh, T.P. | | Deposit date: | 2007-04-23 | | Release date: | 2007-05-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of cloned trimeric hyluranidase from streptococcus pyogenes at 2.8 A resolution
To be Published
|
|
3N2D
 
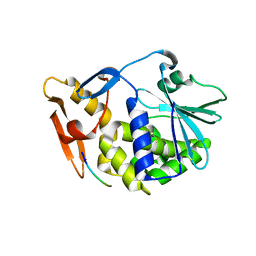 | | Crystal Structure of the Complex of type I Ribosome inactivating protein with hexapeptide Ser-Asp-Asp-Asp-Met-Gly at 2.2 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-meric peptide from 60S acidic ribosomal protein P2-beta, Ribosome inactivating protein | | Authors: | Kushwaha, G.S, Prem Kumar, R, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-05-18 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal Structure of the Complex of type I Ribosome inactivating protein with hexapeptide Ser-Asp-Asp-Asp-Met-Gly at 2.2 A resolution
To be Published
|
|
3NJU
 
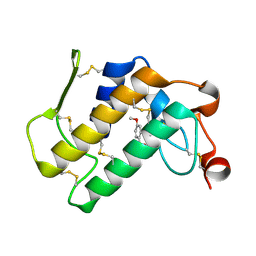 | | Crystal structure of the complex of group I phospholipase A2 with 4-Methoxy-benzoicacid at 1.4A resolution | | Descriptor: | 4-METHOXYBENZOIC ACID, CALCIUM ION, Phospholipase A2 isoform 3 | | Authors: | Kaushik, S, Prem Kumar, R, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-06-18 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the complex of group I phospholipase A2 with 4-Methoxy-benzoicacid at 1.4A resolution
To be Published
|
|
3EKN
 
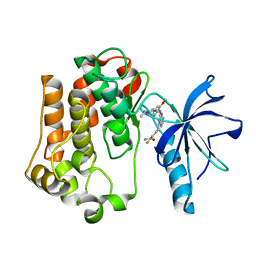 | | Insulin receptor kinase complexed with an inhibitor | | Descriptor: | 2-fluoro-6-{[2-({2-methoxy-4-[4-(1-methylethyl)piperazin-1-yl]phenyl}amino)-7H-pyrrolo[2,3-d]pyrimidin-4-yl]amino}benzamide, Insulin receptor | | Authors: | Chamberlain, S, Atkins, C, Deanda, F, Dumble, M, Gerding, R, Groy, A, Korenchuk, S, Kumar, R, Lei, H, Mook, R, Moorthy, G, Redman, A, Rowland, J, Shewchuk, L. | | Deposit date: | 2008-09-19 | | Release date: | 2008-12-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Optimization of 4,6-bis-anilino-1H-pyrrolo[2,3-d]pyrimidine IGF-1R tyrosine kinase inhibitors towards JNK selectivity.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3EKK
 
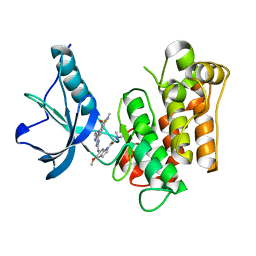 | | Insulin receptor kinase complexed with an inhibitor | | Descriptor: | 2-[(2-{[1-(N,N-dimethylglycyl)-5-methoxy-1H-indol-6-yl]amino}-7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]-6-fluoro-N-methylbenzamide, Insulin receptor | | Authors: | Chamberlain, S, Atkins, C, Deanda, F, Dumble, M, Gerding, R, Groy, A, Korenchuk, S, Kumar, R, Lei, H, Mook, R, Moorthy, G, Redman, A, Rowland, J, Sabbatini, P, Shewchuk, L. | | Deposit date: | 2008-09-19 | | Release date: | 2008-12-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of 4,6-bis-anilino-1H-pyrrolo[2,3-d]pyrimidines: Potent inhibitors of the IGF-1R receptor tyrosine kinase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3ELJ
 
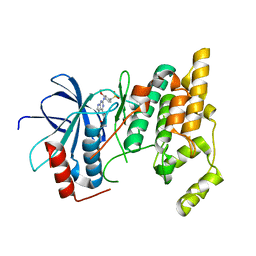 | | Jnk1 complexed with a bis-anilino-pyrrolopyrimidine inhibitor. | | Descriptor: | 2-fluoro-6-{[2-({2-methoxy-4-[(methylsulfonyl)methyl]phenyl}amino)-7H-pyrrolo[2,3-d]pyrimidin-4-yl]amino}benzamide, Mitogen-activated protein kinase 8 | | Authors: | Chamberlain, S, Atkins, C, Deanda, F, Dumble, M, Gerding, R, Groy, A, Korenchuk, S, Kumar, R, Lei, H, Mook, R, Moorthy, G, Redman, A, Rowland, J, Shewchuk, L, Vicentini, G, Mosley, J. | | Deposit date: | 2008-09-22 | | Release date: | 2008-12-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Optimization of 4,6-bis-anilino-1H-pyrrolo[2,3-d]pyrimidine IGF-1R tyrosine kinase inhibitors towards JNK selectivity.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
1SQZ
 
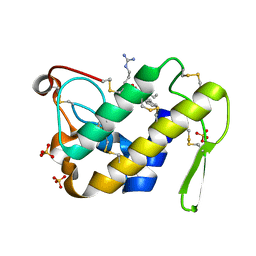 | | Design of specific inhibitors of Phopholipase A2: Crystal structure of the complex formed between Group II Phopholipase A2 and a designed peptide Dehydro-Ile-Ala-Arg-Ser at 1.2A resolution | | Descriptor: | Phospholipase A2, SULFATE ION, synthetic peptide | | Authors: | Singh, N, Prem Kumar, R, Somvanshi, R.K, Bilgrami, S, Ethayathulla, A.S, Sharma, S, Dey, S, Singh, T.P. | | Deposit date: | 2004-03-22 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Design of specific inhibitors of Phopholipase A2: Crystal structure of the complex formed between GroupII Phopholipase A2 and a designed peptide Dehydro-Ile-Ala-Arg-Ser at 1.2A resolution
To be Published
|
|
3ETA
 
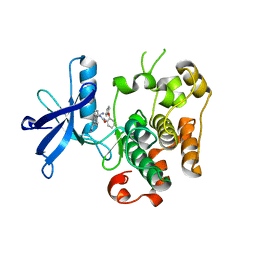 | | Kinase domain of insulin receptor complexed with a pyrrolo pyridine inhibitor | | Descriptor: | 1-(3-{5-[4-(aminomethyl)phenyl]-1H-pyrrolo[2,3-b]pyridin-3-yl}phenyl)-3-(2-phenoxyphenyl)urea, insulin receptor, kinase domain | | Authors: | Patnaik, S, Stevens, K, Gerding, R, Deanda, F, Shotwell, B, Tang, J, Hamajima, T, Nakamura, H, Leesnitzer, A, Hassell, A, Shewchuk, L, Kumar, R, Lei, H, Chamberlain, S. | | Deposit date: | 2008-10-07 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of 3,5-disubstituted-1H-pyrrolo[2,3-b]pyridines as potent inhibitors of the insulin-like growth factor-1 receptor (IGF-1R) tyrosine kinase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4O6S
 
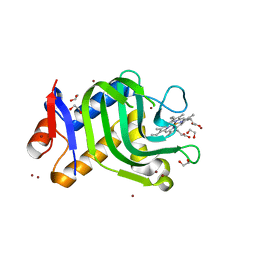 | | 1.32A resolution structure of the hemophore HasA from Pseudomonas aeruginosa (H83A mutant, Zinc Bound) | | Descriptor: | 1,2-ETHANEDIOL, HasAp, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lovell, S, Kumar, R, Battaile, K.P, Matsumura, H, Yao, H, Rodriguez, J.C, Moenne-Loccoz, P, Rivera, M. | | Deposit date: | 2013-12-23 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Replacing the Axial Ligand Tyrosine 75 or Its Hydrogen Bond Partner Histidine 83 Minimally Affects Hemin Acquisition by the Hemophore HasAp from Pseudomonas aeruginosa.
Biochemistry, 53, 2014
|
|
4O6T
 
 | | 1.25A resolution structure of the hemophore HasA from Pseudomonas aeruginosa (H83A mutant, pH 5.4) | | Descriptor: | 1,2-ETHANEDIOL, HasAp, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lovell, S, Kumar, R, Battaile, K.P, Matsumura, H, Yao, H, Rodriguez, J.C, Moenne-Loccoz, P, Rivera, M. | | Deposit date: | 2013-12-23 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Replacing the Axial Ligand Tyrosine 75 or Its Hydrogen Bond Partner Histidine 83 Minimally Affects Hemin Acquisition by the Hemophore HasAp from Pseudomonas aeruginosa.
Biochemistry, 53, 2014
|
|
4O6U
 
 | | 0.89A resolution structure of the hemophore HasA from Pseudomonas aeruginosa (H83A mutant) | | Descriptor: | 1,2-ETHANEDIOL, HasAp, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lovell, S, Kumar, R, Battaile, K.P, Matsumura, H, Yao, H, Rodriguez, J.C, Moenne-Loccoz, P, Rivera, M. | | Deposit date: | 2013-12-23 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Replacing the Axial Ligand Tyrosine 75 or Its Hydrogen Bond Partner Histidine 83 Minimally Affects Hemin Acquisition by the Hemophore HasAp from Pseudomonas aeruginosa.
Biochemistry, 53, 2014
|
|
4O6Q
 
 | | 0.95A resolution structure of the hemophore HasA from Pseudomonas aeruginosa (Y75A mutant) | | Descriptor: | FORMIC ACID, HasAp, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lovell, S, Kumar, R, Battaile, K.P, Matsumura, H, Yao, H, Rodriguez, J.C, Moenne-Loccoz, P, Rivera, M. | | Deposit date: | 2013-12-23 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Replacing the Axial Ligand Tyrosine 75 or Its Hydrogen Bond Partner Histidine 83 Minimally Affects Hemin Acquisition by the Hemophore HasAp from Pseudomonas aeruginosa.
Biochemistry, 53, 2014
|
|
7LXD
 
 | | Structure of yeast DNA Polymerase Zeta (apo) | | Descriptor: | DNA polymerase delta small subunit, DNA polymerase delta subunit 3, DNA polymerase zeta catalytic subunit, ... | | Authors: | Truong, C.D, Craig, T.A, Cui, G, Botuyan, M.V, Serkasevich, R.A, Chan, K.-Y, Mer, G, Chiu, P.-L, Kumar, R. | | Deposit date: | 2021-03-03 | | Release date: | 2021-06-30 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.11 Å) | | Cite: | Cryo-EM reveals conformational flexibility in apo DNA polymerase zeta.
J.Biol.Chem., 297, 2021
|
|
4E6K
 
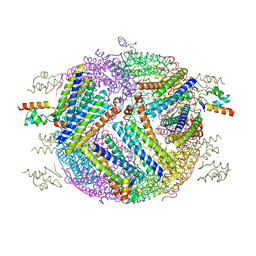 | | 2.0 A resolution structure of Pseudomonas aeruginosa bacterioferritin (BfrB) in complex with bacterioferritin associated ferredoxin (Bfd) | | Descriptor: | Bacterioferritin, FE2/S2 (INORGANIC) CLUSTER, PHOSPHATE ION, ... | | Authors: | Lovell, S, Battaile, K.P, Yao, H, Wang, Y, Kumar, R, Ruvinsky, A, Vasker, I, Rivera, M. | | Deposit date: | 2012-03-15 | | Release date: | 2012-08-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of the BfrB-Bfd Complex Reveals Protein-Protein Interactions Enabling Iron Release from Bacterioferritin.
J.Am.Chem.Soc., 134, 2012
|
|
3RUV
 
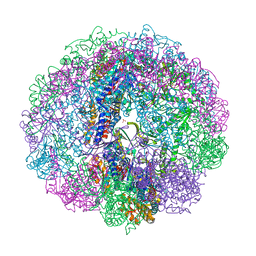 | | Crystal structure of Cpn-rls in complex with ATP analogue from Methanococcus maripaludis | | Descriptor: | Chaperonin, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Pereira, J.H, Ralston, C.Y, Douglas, N.R, Kumar, R, McAndrew, R.P, Knee, K.M, King, J.A, Frydman, J, Adams, P.D. | | Deposit date: | 2011-05-05 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.242 Å) | | Cite: | Mechanism of nucleotide sensing in group II chaperonins.
Embo J., 31, 2012
|
|
3RUQ
 
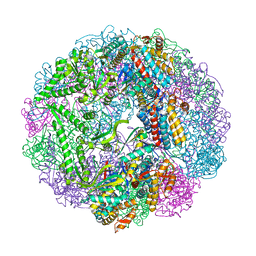 | | Crystal structure of Cpn-WT in complex with ADP from Methanococcus maripaludis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperonin, MAGNESIUM ION | | Authors: | Pereira, J.H, Ralston, C.Y, Douglas, N.R, Kumar, R, McAndrew, R.P, Knee, K.M, Frydman, J, Adams, P.D. | | Deposit date: | 2011-05-05 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | Mechanism of nucleotide sensing in group II chaperonins.
Embo J., 31, 2012
|
|
3RUS
 
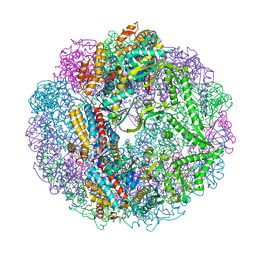 | | Crystal structure of Cpn-rls in complex with ADP from Methanococcus maripaludis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperonin, MAGNESIUM ION, ... | | Authors: | Pereira, J.H, Ralston, C.Y, Douglas, N.R, Kumar, R, McAndrew, R.P, Knee, K.M, King, J.A, Frydman, J, Adams, P.D. | | Deposit date: | 2011-05-05 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.338 Å) | | Cite: | Mechanism of nucleotide sensing in group II chaperonins.
Embo J., 31, 2012
|
|
3RUW
 
 | | Crystal structure of Cpn-rls in complex with ADP-AlFx from Methanococcus maripaludis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Chaperonin, ... | | Authors: | Pereira, J.H, Ralston, C.Y, Douglas, N.R, Kumar, R, McAndrew, R.P, Knee, K.M, King, J.A, Frydman, J, Adams, P.D. | | Deposit date: | 2011-05-05 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Mechanism of nucleotide sensing in group II chaperonins.
Embo J., 31, 2012
|
|
2E9E
 
 | | Crystal structure of the complex of goat lactoperoxidase with Nitrate at 3.25 A resolution | | Descriptor: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Singh, A.K, Prem kumar, R, Singh, N, Sharma, S, Singh, S.B, Bhushan, A, Kaur, P, Singh, T.P. | | Deposit date: | 2007-01-25 | | Release date: | 2007-02-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structure of the complex of goat lactoperoxidase with Nitrate at 3.25 A resolution
To be Published
|
|
1ZWP
 
 | | The atomic resolution Crystal structure of the Phospholipase A2 (PLA2) complex with Nimesulide reveals its weaker binding to PLA2 | | Descriptor: | 4-NITRO-2-PHENOXYMETHANESULFONANILIDE, METHANOL, Phospholipase A2 VRV-PL-VIIIa, ... | | Authors: | Prem Kumar, R, Singh, N, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2005-06-04 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The atomic resolution Crystal structure of the Phospholipase A2 (PLA2) complex with Nimesulide reveals its weaker binding to PLA2
To be Published
|
|
2F33
 
 | | NMR solution structure of Ca2+-loaded calbindin D28K | | Descriptor: | Calbindin | | Authors: | Kojetin, D.J, Venters, R.A, Kordys, D.R, Thompson, R.J, Kumar, R, Cavanagh, J. | | Deposit date: | 2005-11-18 | | Release date: | 2006-07-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure, binding interface and hydrophobic transitions of Ca(2+)-loaded calbindin-D(28K).
Nat.Struct.Mol.Biol., 13, 2006
|
|
2HD4
 
 | | Crystal structure of proteinase K inhibited by a lactoferrin octapeptide Gly-Asp-Glu-Gln-Gly-Glu-Asn-Lys at 2.15 A resolution | | Descriptor: | 8-mer Peptide from Lactotransferrin, ACETIC ACID, CALCIUM ION, ... | | Authors: | Prem Kumar, R, Singh, A.K, Singh, N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2006-06-20 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of proteinase K inhibited by a lactoferrin octapeptide Gly-Asp-Glu-Gln-Gly-Glu-Asn-Lys at 2.15 A resolution
To be Published
|
|
2HPZ
 
 | | Crystal structure of proteinase K complex with a synthetic peptide KLKLLVVIRLK at 1.69 A resolution | | Descriptor: | 11-mer synthetic peptide, CALCIUM ION, NITRATE ION, ... | | Authors: | Prem kumar, R, Singh, A.K, Somvanshi, R.K, Singh, N, Sharma, S, Kaur, P, Dey, S, Bhushan, A, Singh, T.P. | | Deposit date: | 2006-07-18 | | Release date: | 2006-08-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of proteinase K complex with a synthetic peptide KLKLLVVIRLK at 1.69 A resolution
To be Published
|
|
2G58
 
 | | Crystal structure of a complex of phospholipase A2 with a designed peptide inhibitor Dehydro-Ile-Ala-Arg-Ser at 0.98 A resolution | | Descriptor: | (PHQ)IARS, Phospholipase A2 VRV-PL-VIIIa, SULFATE ION | | Authors: | Prem Kumar, R, Singh, N, Somvanshi, R.K, Ethayathulla, A.S, Dey, S, Sharma, S, Kaur, P, Perbandt, M, Betzel, C, Singh, T.P. | | Deposit date: | 2006-02-22 | | Release date: | 2006-03-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Crystal structure of a complex of phospholipase A2 with a designed peptide inhibitor Dehydro-Ile-Ala-Arg-Ser at 0.98 A resolution
To be Published
|
|
2G9B
 
 | | NMR solution structure of CA2+-loaded calbindin D28K | | Descriptor: | Calbindin | | Authors: | Kojetin, D.J, Venters, R.A, Kordys, D.R, Thompson, R.J, Kumar, R, Cavanagh, J. | | Deposit date: | 2006-03-06 | | Release date: | 2006-07-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure, binding interface and hydrophobic transitions of Ca(2+)-loaded calbindin-D(28K).
Nat.Struct.Mol.Biol., 13, 2006
|
|
