6NP2
 
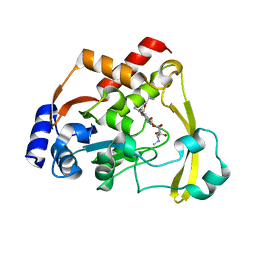 | | AAC-VIa bound to Sisomicin | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, Aminoglycoside N(3)-acetyltransferase, MAGNESIUM ION | | Authors: | Kumar, P, Cuneo, M.J. | | Deposit date: | 2019-01-17 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Low-Barrier and Canonical Hydrogen Bonds Modulate Activity and Specificity of a Catalytic Triad.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
5WXU
 
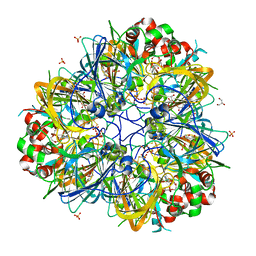 | | 11S globulin from Wrightia tinctoria reveals auxin binding site | | Descriptor: | 11S globulin, 1H-INDOL-3-YLACETIC ACID, CITRATE ANION, ... | | Authors: | Kumar, P, Kesari, P, Dhindwal, S, Kumar, P. | | Deposit date: | 2017-01-09 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A novel function for globulin in sequestering plant hormone: Crystal structure of Wrightia tinctoria 11S globulin in complex with auxin.
Sci Rep, 7, 2017
|
|
4R62
 
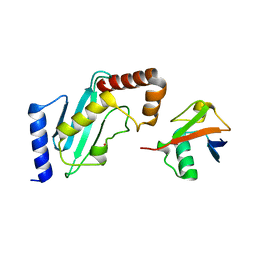 | | Structure of Rad6~Ub | | Descriptor: | ACETATE ION, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-conjugating enzyme E2 2 | | Authors: | Kumar, P, Wolberger, C. | | Deposit date: | 2014-08-22 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Role of a non-canonical surface of Rad6 in ubiquitin conjugating activity.
Nucleic Acids Res., 43, 2015
|
|
1I6B
 
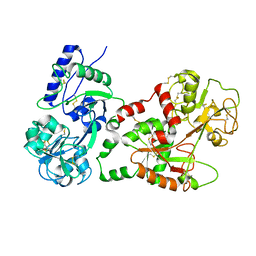 | |
3GZY
 
 | | Crystal Structure of the Biphenyl Dioxygenase from Comamonas testosteroni Sp. Strain B-356 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Biphenyl dioxygenase subunit alpha, Biphenyl dioxygenase subunit beta, ... | | Authors: | Kumar, P, Colbert, C.L, Bolin, J.T. | | Deposit date: | 2009-04-08 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural Characterization of Pandoraea pnomenusa B-356 Biphenyl Dioxygenase Reveals Features of Potent Polychlorinated Biphenyl-Degrading Enzymes
Plos One, 8, 2013
|
|
8BFD
 
 | | Racemic structure of PK-7 (310HD-U2U5) | | Descriptor: | 310HD-U2U5, D-310HD-U2U5, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kumar, P, Paterson, N.G, Woolfson, D.N. | | Deposit date: | 2022-10-25 | | Release date: | 2022-11-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | De novo design of discrete, stable 3 10 -helix peptide assemblies.
Nature, 607, 2022
|
|
8BFE
 
 | | A dimeric de novo coiled-coil assembly: PK-2 (CC-TypeN-LaUbUcLd) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, CC-TypeN-LaUbUcLd, ... | | Authors: | Kumar, P, Paterson, N.G, Woolfson, D.N. | | Deposit date: | 2022-10-25 | | Release date: | 2022-11-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | De novo design of discrete, stable 3 10 -helix peptide assemblies.
Nature, 607, 2022
|
|
3H3U
 
 | | Crystal structure of CRP (cAMP receptor Protein) from Mycobacterium tuberculosis | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN (PROBABLY CRP/FNR-FAMILY), SULFATE ION | | Authors: | Kumar, P, Joshi, D.C, Akif, M, Akhter, Y, Hasnain, S.E, Mande, S.C. | | Deposit date: | 2009-04-17 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mapping conformational transitions in cyclic AMP receptor protein: crystal structure and normal-mode analysis of Mycobacterium tuberculosis apo-cAMP receptor protein
Biophys.J., 98, 2010
|
|
1JW1
 
 | |
8B45
 
 | | Structure of CC-Tri with Aib@b,c: CC-Tri-(UbUc)4 | | Descriptor: | 1,2-ETHANEDIOL, CC-Tri-(UbUc)4, SODIUM ION, ... | | Authors: | Kumar, P, Martin, F.J.O, Dawson, W.M, Zieleniewski, F, Woolfson, D.N. | | Deposit date: | 2022-09-19 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of CC-Tri with Aib@b,c: CC-Tri-(UbUc)4
To Be Published
|
|
3BVN
 
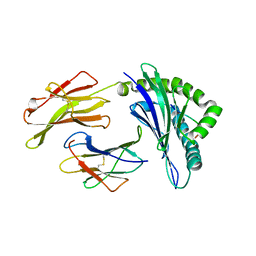 | | High resolution crystal structure of HLA-B*1402 in complex with the latent membrane protein 2 peptide (LMP2) of Epstein-Barr virus | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B*1402 alpha chain, ... | | Authors: | Kumar, P, Vahedi-Faridi, A, Saenger, W, Uchanska-Ziegler, B, Ziegler, A. | | Deposit date: | 2008-01-07 | | Release date: | 2009-02-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis for T cell alloreactivity among three HLA-B14 and HLA-B27 antigens
J.Biol.Chem., 284, 2009
|
|
3LQU
 
 | |
3LRJ
 
 | |
3NQ4
 
 | |
3MK3
 
 | | Crystal structure of Lumazine synthase from Salmonella typhimurium LT2 | | Descriptor: | 6,7-dimethyl-8-ribityllumazine synthase, SULFATE ION | | Authors: | Kumar, P, Singh, M, Karthikeyan, S. | | Deposit date: | 2010-04-14 | | Release date: | 2011-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.569 Å) | | Cite: | Crystal structure analysis of icosahedral lumazine synthase from Salmonella typhimurium, an antibacterial drug target.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
5DUJ
 
 | | Crystal structure of ldtMt2 in complex with Faropenem adduct | | Descriptor: | (2R,5R)-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-5-[(2R)-tetrahydrofuran-2-yl]-2,5-dihydro-1,3-thiazole-4-carboxylic acid, BETA-MERCAPTOETHANOL, L,D-transpeptidase 2, ... | | Authors: | Kumar, P, Lamichhane, G. | | Deposit date: | 2015-09-18 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Non-classical transpeptidases yield insight into new antibacterials.
Nat. Chem. Biol., 13, 2017
|
|
5DU7
 
 | |
5E1I
 
 | | Crystal structure of Mycobacterium tuberculosis L,D-transpeptidase 2 with carbapenem drug T210 | | Descriptor: | (2S,3R,4R)-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-4-(methylsulfanyl)-3,4-dihydro-2H-pyrrole-5-carboxylic acid, GLYCEROL, L,D-transpeptidase 2, ... | | Authors: | Kumar, P, Ginell, S.L, Lamichhane, G. | | Deposit date: | 2015-09-29 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Non-classical transpeptidases yield insight into new antibacterials.
Nat. Chem. Biol., 13, 2017
|
|
5DVP
 
 | | Crystal structure of Mycobacterium tuberculosis L,D-transpeptidase 2 with Doripenem adduct | | Descriptor: | (2S,3R,4S)-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-4-({(3S,5S)-5-[(sulfamoylamino)methyl]pyrrolidin-3-yl}sulfanyl)-3,4-dihydro-2H-pyrrole-5-carboxylic acid, L,D-transpeptidase 2, PHOSPHONOACETALDEHYDE, ... | | Authors: | Kumar, P, Lamichhane, G. | | Deposit date: | 2015-09-21 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Non-classical transpeptidases yield insight into new antibacterials.
Nat. Chem. Biol., 13, 2017
|
|
5E1G
 
 | | Crystal structure of Mycobacterium tuberculosis L,D-transpeptidase 2 with carbapenem drug T208 | | Descriptor: | (2~{S},3~{R},4~{R})-4-(2-cyclohexyloxy-2-oxidanylidene-ethyl)sulfanyl-3-methyl-2-[(2~{S},3~{R})-3-oxidanyl-1-oxidanylidene-butan-2-yl]-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, L,D-transpeptidase 2 | | Authors: | Kumar, P, Lamichhane, G, Ginell, S.L. | | Deposit date: | 2015-09-29 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Non-classical transpeptidases yield insight into new antibacterials.
Nat. Chem. Biol., 13, 2017
|
|
5DZP
 
 | | Crystal structure of Mycobacterium tuberculosis L,D-transpeptidase 2 with carbapenem drug T206 in conformation B | | Descriptor: | (2~{R},3~{R},4~{R})-4-methyl-3-(2-oxidanylidene-2-propoxy-ethyl)sulfanyl-5-[(2~{S},3~{R})-3-oxidanyl-1-oxidanylidene-butan-2-yl]-3,4-dihydro-2~{H}-pyrrole-2-carboxylic acid, L,D-transpeptidase 2 | | Authors: | Kumar, P, Ginell, S.L, Lamichhane, G. | | Deposit date: | 2015-09-25 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Non-classical transpeptidases yield insight into new antibacterials.
Nat. Chem. Biol., 13, 2017
|
|
7L6U
 
 | |
5E5L
 
 | |
5E51
 
 | | Crystal structure of Mycobacterium tuberculosis L,D-transpeptidase 1 with Faropenem adduct | | Descriptor: | (3R)-3-hydroxybutanal, L,D-transpeptidase 1 | | Authors: | Kumar, P, Lamichhane, G, Ginell, S.L. | | Deposit date: | 2015-10-07 | | Release date: | 2016-10-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Non-classical transpeptidases yield insight into new antibacterials.
Nat. Chem. Biol., 13, 2017
|
|
7K34
 
 | |
