4AIB
 
 | |
8JFV
 
 | | Crystal structure of Catabolite repressor acivator from E. coli in complex with sulisobenzone | | Descriptor: | 1,2-ETHANEDIOL, 2-methoxy-4-oxidanyl-5-(phenylcarbonyl)benzenesulfonic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Neetu, N, Sharma, M, Mahto, J.K, Kumar, P. | | Deposit date: | 2023-05-19 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Sulisobenzone is a potent inhibitor of the global transcription factor Cra.
J.Struct.Biol., 215, 2023
|
|
8JFF
 
 | |
4B16
 
 | |
4B15
 
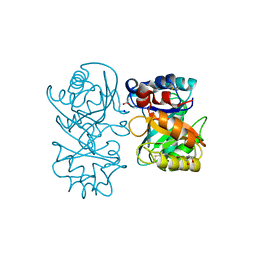 | | crystal structure of tamarind chitinase like lectin (TCLL) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Patil, D.N, Kumar, P. | | Deposit date: | 2012-07-06 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural Investigation of a Novel N-Acetyl Glucosamine Binding Chi-Lectin which Reveals Evolutionary Relationship with Class III Chitinases.
Plos One, 8, 2013
|
|
5K69
 
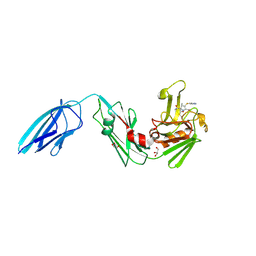 | | Crystal structure of Mycobacterium tuberculosis L,D-transpeptidase 2 with carbapenem drug T224 | | Descriptor: | (2~{S},3~{R},4~{R})-4-(1~{H}-indol-3-ylsulfanyl)-3-methyl-2-[(2~{S},3~{S})-3-oxidanyl-1-oxidanylidene-butan-2-yl]-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, GLYCEROL, L,D-transpeptidase 2 | | Authors: | Lamichhane, G, Ginell, S.L, Kumar, P. | | Deposit date: | 2016-05-24 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Non-classical transpeptidases yield insight into new antibacterials.
Nat. Chem. Biol., 13, 2017
|
|
6LK2
 
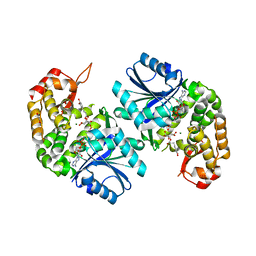 | | Crystal structure of Providencia alcalifaciens 3-dehydroquinate synthase (DHQS) in complex with Mg2+, NAD and chlorogenic acid | | Descriptor: | (1R,3R,4S,5R)-3-[3-[3,4-bis(oxidanyl)phenyl]propanoyloxy]-1,4,5-tris(oxidanyl)cyclohexane-1-carboxylic acid, 1,2-ETHANEDIOL, 3-dehydroquinate synthase, ... | | Authors: | Neetu, N, Katiki, M, Kumar, P. | | Deposit date: | 2019-12-17 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Structural and Biochemical Analyses Reveal that Chlorogenic Acid Inhibits the Shikimate Pathway.
J.Bacteriol., 202, 2020
|
|
7T7L
 
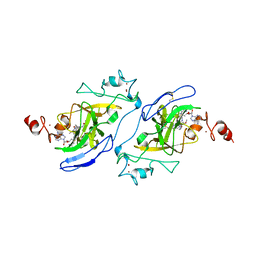 | | Structure of human G9a SET-domain (EHMT2) in complex with covalent inhibitor (Compound 1) | | Descriptor: | Histone-lysine N-methyltransferase EHMT2, N-(6-methoxy-4-{[1-(propan-2-yl)piperidin-4-yl]amino}-7-[3-(pyrrolidin-1-yl)propoxy]quinazolin-2-yl)prop-2-enamide, N-(6-methoxy-4-{[1-(propan-2-yl)piperidin-4-yl]amino}-7-[3-(pyrrolidin-1-yl)propoxy]quinazolin-2-yl)propanamide, ... | | Authors: | Park, K.-S, Kumar, P. | | Deposit date: | 2021-12-15 | | Release date: | 2022-07-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of the First-in-Class G9a/GLP Covalent Inhibitors.
J.Med.Chem., 65, 2022
|
|
7T7M
 
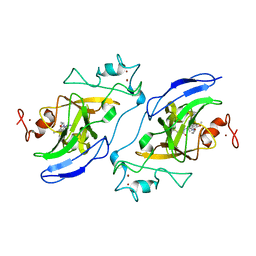 | | Structure of human GLP SET-domain (EHMT1) in complex with covalent inhibitor (Compound 1) | | Descriptor: | Histone-lysine N-methyltransferase EHMT1, N-(6-methoxy-4-{[1-(propan-2-yl)piperidin-4-yl]amino}-7-[3-(pyrrolidin-1-yl)propoxy]quinazolin-2-yl)prop-2-enamide, N-(6-methoxy-4-{[1-(propan-2-yl)piperidin-4-yl]amino}-7-[3-(pyrrolidin-1-yl)propoxy]quinazolin-2-yl)propanamide, ... | | Authors: | Park, K.-S, Kumar, P. | | Deposit date: | 2021-12-15 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery of the First-in-Class G9a/GLP Covalent Inhibitors.
J.Med.Chem., 65, 2022
|
|
1IC6
 
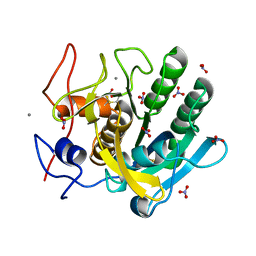 | | STRUCTURE OF A SERINE PROTEASE PROTEINASE K FROM TRITIRACHIUM ALBUM LIMBER AT 0.98 A RESOLUTION | | Descriptor: | CALCIUM ION, NITRATE ION, PROTEINASE K | | Authors: | Betzel, C, Gourinath, S, Kumar, P, Kaur, P, Perbandt, M, Eschenburg, S, Singh, T.P. | | Deposit date: | 2001-03-30 | | Release date: | 2001-04-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Structure of a serine protease proteinase K from Tritirachium album limber at 0.98 A resolution.
Biochemistry, 40, 2001
|
|
7V25
 
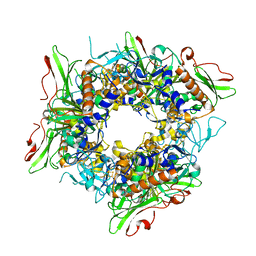 | |
7V28
 
 | |
1LJY
 
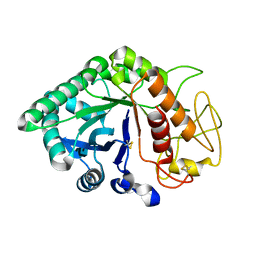 | | Crystal Structure of a Novel Regulatory 40 kDa Mammary Gland Protein (MGP-40) secreted during Involution | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MGP-40 | | Authors: | Mohanty, A.K, Singh, G, Paramasivam, M, Saravanan, K, Jabeen, T, Sharma, S, Yadav, S, Kaur, P, Kumar, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2002-04-23 | | Release date: | 2003-03-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of a Novel Regulatory 40 kDa Mammary Gland Protein (MGP-40) secreted during Involution
J.Biol.Chem., 278, 2003
|
|
5GMU
 
 | | Crystal structure of chorismate mutase like domain of bifunctional DAHP synthase of Bacillus subtilis in complex with Chlorogenic acid | | Descriptor: | (1R,3R,4S,5R)-3-[3-[3,4-bis(oxidanyl)phenyl]propanoyloxy]-1,4,5-tris(oxidanyl)cyclohexane-1-carboxylic acid, Protein AroA(G), SULFATE ION | | Authors: | Pratap, S, Dev, A, Sharma, V, Yadav, R, Narwal, M, Tomar, S, Kumar, P. | | Deposit date: | 2016-07-16 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Chorismate Mutase-like Domain of DAHPS from Bacillus subtilis Complexed with Novel Inhibitor Reveals Conformational Plasticity of Active Site.
Sci Rep, 7, 2017
|
|
5GO2
 
 | | Crystal structure of chorismate mutase like domain of bifunctional DAHP synthase of Bacillus subtilis in complex with Citrate | | Descriptor: | CITRIC ACID, Protein AroA(G), SULFATE ION | | Authors: | Pratap, S, Dev, A, Sharma, V, Yadav, R, Narwal, M, Tomar, S, Kumar, P. | | Deposit date: | 2016-07-26 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.907 Å) | | Cite: | Structure of Chorismate Mutase-like Domain of DAHPS from Bacillus subtilis Complexed with Novel Inhibitor Reveals Conformational Plasticity of Active Site.
Sci Rep, 7, 2017
|
|
7X1X
 
 | | Crystal Structure of cis-4,5-dihydrodiol phthalate dehydrogenase in complex with NAD+ | | Descriptor: | 4,5-dihydroxyphthalate dehydrogenase, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Sharma, M, Mahto, J.K, Kumar, P. | | Deposit date: | 2022-02-24 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Conformational flexibility enables catalysis of phthalate cis-4,5-dihydrodiol dehydrogenase.
Arch.Biochem.Biophys., 727, 2022
|
|
7X2Y
 
 | | Crystal Structure of cis-4,5-dihydrodiol phthalate dehydrogenase in complex with NAD+ and 3-Hydroxybenzoate | | Descriptor: | 3-HYDROXYBENZOIC ACID, 4,5-dihydroxyphthalate dehydrogenase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sharma, M, Mahto, J.K, Kumar, P. | | Deposit date: | 2022-02-26 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Conformational flexibility enables catalysis of phthalate cis-4,5-dihydrodiol dehydrogenase.
Arch.Biochem.Biophys., 727, 2022
|
|
7WZD
 
 | | Crystal Structure of cis-4,5-dihydrodiol phthalate dehydrogenase from Comamonas testosteroni KF1 | | Descriptor: | 4,5-dihydroxyphthalate dehydrogenase, GLYCEROL | | Authors: | Sharma, M, Mahto, J.K, Kumar, P. | | Deposit date: | 2022-02-17 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational flexibility enables catalysis of phthalate cis-4,5-dihydrodiol dehydrogenase.
Arch.Biochem.Biophys., 727, 2022
|
|
7VJU
 
 | |
5XLW
 
 | | Mycobacterium tuberculosis Pantothenate kinase mutant F247A/F254A | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, ... | | Authors: | Paul, A, Kumar, P, Surolia, A, Vijayan, M. | | Deposit date: | 2017-05-11 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Biochemical and structural studies of mutants indicate concerted movement of the dimer interface and ligand-binding region of Mycobacterium tuberculosis pantothenate kinase
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
3VB0
 
 | | Crystal structure of 2,2',3-trihydroxybiphenyl 1,2-dioxygenase from dibenzofuran-degrading Sphingomonas wittichii strain RW1 | | Descriptor: | FE (II) ION, Glyoxalase/bleomycin resistance protein/dioxygenase, HEXAETHYLENE GLYCOL, ... | | Authors: | Koksal, M, Kumar, P, Bolin, J.T. | | Deposit date: | 2011-12-30 | | Release date: | 2013-04-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a dibenzofuran-degrading dioxygenase: an unusual spatially heterogeneous crystal with a hypersymmetric intensity distribution
To be Published
|
|
5XMB
 
 | | Mycobacterium tuberculosis Pantothenate kinase mutant F247A | | Descriptor: | Pantothenate kinase, SULFATE ION | | Authors: | Paul, A, Kumar, P, Surolia, A, Vijayan, M. | | Deposit date: | 2017-05-13 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Biochemical and structural studies of mutants indicate concerted movement of the dimer interface and ligand-binding region of Mycobacterium tuberculosis pantothenate kinase
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
3ZHW
 
 | | X-ray Crystallographic Structural Characteristics of Arabidopsis Hemoglobin I and their Functional Implications | | Descriptor: | NON-SYMBIOTIC HEMOGLOBIN 1, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Mukhi, N, Dhindwal, S, Uppal, S, Kumar, P, Kaur, J, Kundu, S. | | Deposit date: | 2012-12-30 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | X-Ray Crystallographic Structural Characteristics of Arabidopsis Hemoglobin I and Their Functional Implications
Biochim.Biophys.Acta, 1834, 2013
|
|
3ZQ7
 
 | |
5XLV
 
 | | Mycobacterium tuberculosis Pantothenate kinase mutant F254A | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Pantothenate kinase, ... | | Authors: | Paul, A, Kumar, P, Surolia, A, Vijayan, M. | | Deposit date: | 2017-05-11 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical and structural studies of mutants indicate concerted movement of the dimer interface and ligand-binding region of Mycobacterium tuberculosis pantothenate kinase
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
