5DL2
 
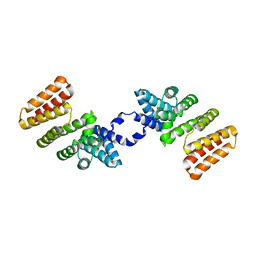 | | Crystal Structure of RopB | | Descriptor: | Regulator of protease B (RopB) | | Authors: | Kumaraswami, M. | | Deposit date: | 2015-09-04 | | Release date: | 2016-01-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural and functional analysis of RopB: a major virulence regulator in Streptococcus pyogenes.
Mol.Microbiol., 99, 2016
|
|
1RR7
 
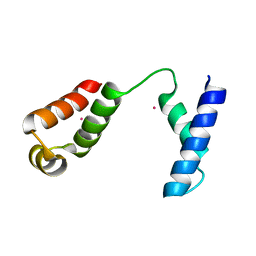 | |
1EHS
 
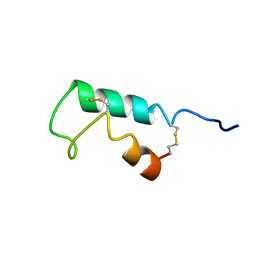 | | THE STRUCTURE OF ESCHERICHIA COLI HEAT-STABLE ENTEROTOXIN B BY NUCLEAR MAGNETIC RESONANCE AND CIRCULAR DICHROISM | | Descriptor: | HEAT-STABLE ENTEROTOXIN B | | Authors: | Sukumar, M, Rizo, J, Wall, M, Dreyfus, L.A, Kupersztoch, Y.M, Gierasch, L.M. | | Deposit date: | 1995-06-13 | | Release date: | 1995-09-15 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The structure of Escherichia coli heat-stable enterotoxin b by nuclear magnetic resonance and circular dichroism.
Protein Sci., 4, 1995
|
|
3VIB
 
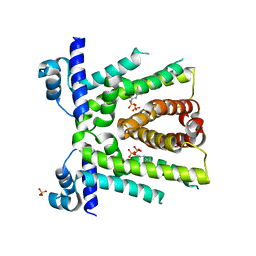 | |
4GPA
 
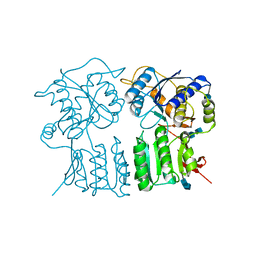 | | High resolution structure of the GluA4 N-terminal domain (NTD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 4 | | Authors: | Sukumaran, M, Greger, I.H. | | Deposit date: | 2012-08-20 | | Release date: | 2012-10-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Comparative Dynamics of NMDA- and AMPA-Glutamate Receptor N-Terminal Domains.
Structure, 20, 2012
|
|
3IAO
 
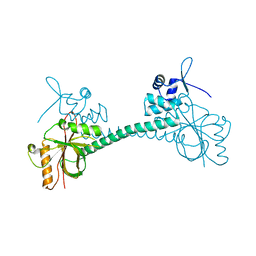 | |
3BM5
 
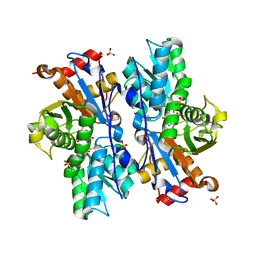 | | Crystal structure of O-acetyl-serine sulfhydrylase from Entamoeba histolytica in complex with cysteine | | Descriptor: | CYSTEINE, Cysteine synthase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Krishna, C, Kumar, M, Kumar, S, Gourinath, S. | | Deposit date: | 2007-12-12 | | Release date: | 2008-04-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of native O-acetyl-serine sulfhydrylase from Entamoeba histolytica and its complex with cysteine: structural evidence for cysteine binding and lack of interactions with serine acetyl transferase.
Proteins, 72, 2008
|
|
6TXN
 
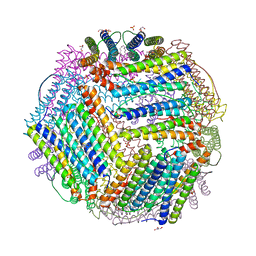 | | Crystal structure of thermotoga maritima Ferritin in apo form | | Descriptor: | EICOSANE, Ferritin, GLYCEROL, ... | | Authors: | Wilk, P, Grudnik, P, Kumar, M, Heddle, J, Chakraborti, S. | | Deposit date: | 2020-01-14 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A single residue can modulate nanocage assembly in salt dependent ferritin.
Nanoscale, 13, 2021
|
|
6TXM
 
 | | Crystal structure of thermotoga maritima E65R Ferritin | | Descriptor: | EICOSANE, Ferritin, GLYCEROL, ... | | Authors: | Wilk, P, Grudnik, P, Kumar, M, Heddle, J, Chakraborti, S. | | Deposit date: | 2020-01-14 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | A single residue can modulate nanocage assembly in salt dependent ferritin.
Nanoscale, 13, 2021
|
|
6TXH
 
 | | Crystal structure of thermotoga maritima Ferritin in apo form | | Descriptor: | EICOSANE, Ferritin, GLYCEROL, ... | | Authors: | Wilk, P, Grudnik, P, Kumar, M, Heddle, J, Chakraborti, S. | | Deposit date: | 2020-01-14 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | A single residue can modulate nanocage assembly in salt dependent ferritin.
Nanoscale, 13, 2021
|
|
6TXJ
 
 | | Crystal structure of thermotoga maritima A42V E65D Ferritin | | Descriptor: | EICOSANE, FE (III) ION, Ferritin, ... | | Authors: | Wilk, P, Grudnik, P, Kumar, M, Heddle, J, Chakraborti, S, Biela, A.P. | | Deposit date: | 2020-01-14 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | A single residue can modulate nanocage assembly in salt dependent ferritin.
Nanoscale, 13, 2021
|
|
6TXL
 
 | | Crystal structure of thermotoga maritima E65Q Ferritin | | Descriptor: | EICOSANE, FE (III) ION, Ferritin, ... | | Authors: | Wilk, P, Grudnik, P, Kumar, M, Heddle, J, Chakraborti, S. | | Deposit date: | 2020-01-14 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | A single residue can modulate nanocage assembly in salt dependent ferritin.
Nanoscale, 13, 2021
|
|
6TXK
 
 | | Crystal structure of thermotoga maritima E65K Ferritin | | Descriptor: | EICOSANE, FE (III) ION, Ferritin, ... | | Authors: | Wilk, P, Grudnik, P, Kumar, M, Heddle, J, Chakraborti, S. | | Deposit date: | 2020-01-14 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.359 Å) | | Cite: | A single residue can modulate nanocage assembly in salt dependent ferritin.
Nanoscale, 13, 2021
|
|
6TXI
 
 | | Crystal structure of thermotoga maritima E65A Ferritin | | Descriptor: | EICOSANE, FE (III) ION, Ferritin, ... | | Authors: | Wilk, P, Grudnik, P, Kumar, M, Heddle, J, Chakraborti, S. | | Deposit date: | 2020-01-14 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.759 Å) | | Cite: | A single residue can modulate nanocage assembly in salt dependent ferritin.
Nanoscale, 13, 2021
|
|
7ZUG
 
 | | Heterogeneous nuclear ribonucleoprotein H1, qRRM2 domain | | Descriptor: | CHLORIDE ION, Heterogeneous nuclear ribonucleoprotein H, N-terminally processed, ... | | Authors: | Winter, N, Kumar, M, Isupov, M.N, Wiener, R. | | Deposit date: | 2022-05-12 | | Release date: | 2023-05-24 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.075 Å) | | Cite: | Heterogeneous nuclear ribonucleoprotein H1, qRRM2 domain
To Be Published
|
|
8AXG
 
 | | Crystal structure of Fusobacterium nucleatum fusolisin protease | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Fusolisin, ... | | Authors: | Isupov, M.N, Wiener, R, Rouvinski, A, Fahoum, J, Kumar, M, Read, R.J. | | Deposit date: | 2022-08-31 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of Fusobacterium nucleatum fusolisin protease
To Be Published
|
|
4RC9
 
 | | Crystal Structure of the type II Dehydroquinate dehydratase from Acinetobacter baumannii at 2.03A Resolution | | Descriptor: | 3-dehydroquinate dehydratase, SULFATE ION | | Authors: | Iqbal, N, Kumar, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-09-15 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of the type II Dehydroquinate dehydratase from Acinetobacter baumannii at 2.03 A Resolution
To be Published
|
|
7QUT
 
 | | serial synchrotron crystallographic structure of Drosophila Melanogaster (6-4) photolyase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, RE11660p | | Authors: | Cellini, A, Weixiao, Y.W, Kumar, M.S, Westenhoff, S. | | Deposit date: | 2022-01-18 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural basis of the radical pair state in photolyases and cryptochromes.
Chem.Commun.(Camb.), 58, 2022
|
|
6KNO
 
 | |
6KMY
 
 | | Structure of single disulfide peptide Czon1107-P5A | | Descriptor: | Czon1107-P5A | | Authors: | Sarma, S.P, Madhan Kumar, M. | | Deposit date: | 2019-08-01 | | Release date: | 2020-04-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and allosteric activity of a single-disulfide conopeptide fromConus zonatusat human alpha 3 beta 4 and alpha 7 nicotinic acetylcholine receptors.
J.Biol.Chem., 295, 2020
|
|
6KNP
 
 | |
6KN2
 
 | |
6KN3
 
 | |
3PUO
 
 | | Crystal structure of dihydrodipicolinate synthase from Pseudomonas aeruginosa(PsDHDPS)complexed with L-lysine at 2.65A resolution | | Descriptor: | Dihydrodipicolinate synthase, GLYCEROL, LYSINE | | Authors: | Kaur, N, Kumar, M, Kumar, S, Gautam, A, Sinha, M, Kaur, P, Sharma, S, Sharma, R, Tewari, R, Singh, T.P. | | Deposit date: | 2010-12-06 | | Release date: | 2010-12-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Biochemical studies and crystal structure determination of dihydrodipicolinate synthase from Pseudomonas aeruginosa
Int.J.Biol.Macromol., 48, 2011
|
|
8C6C
 
 | | Light SFX structure of D.m(6-4)photolyase at 300ps time delay | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
