5T77
 
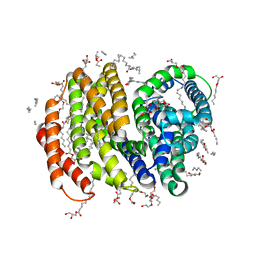 | | Crystal structure of the MOP flippase MurJ | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CALCIUM ION, ... | | Authors: | Kuk, A.C.Y, Lee, S.-Y. | | Deposit date: | 2016-09-02 | | Release date: | 2016-12-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the MOP flippase MurJ in an inward-facing conformation.
Nat. Struct. Mol. Biol., 24, 2017
|
|
6NC6
 
 | | Lipid II flippase MurJ, inward closed conformation | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Lipid II flippase MurJ, ... | | Authors: | Kuk, A.C.Y, Lee, S.-Y. | | Deposit date: | 2018-12-10 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Visualizing conformation transitions of the Lipid II flippase MurJ.
Nat Commun, 10, 2019
|
|
6NC7
 
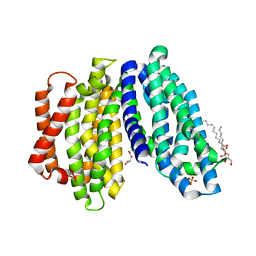 | | Lipid II flippase MurJ, inward open conformation | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Lipid II flippase MurJ, SULFATE ION | | Authors: | Kuk, A.C.Y, Lee, S.-Y. | | Deposit date: | 2018-12-11 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Visualizing conformation transitions of the Lipid II flippase MurJ.
Nat Commun, 10, 2019
|
|
6NC8
 
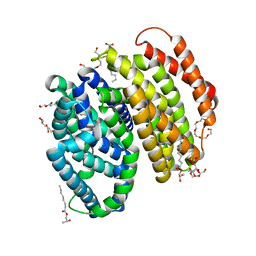 | | Lipid II flippase MurJ, inward occluded conformation | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Lipid II flippase MurJ, PENTAETHYLENE GLYCOL, ... | | Authors: | Kuk, A.C.Y, Lee, S.-Y. | | Deposit date: | 2018-12-11 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Visualizing conformation transitions of the Lipid II flippase MurJ.
Nat Commun, 10, 2019
|
|
6NC9
 
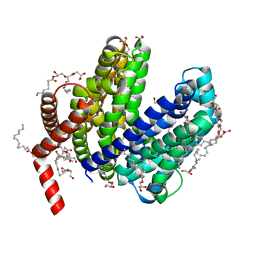 | | Lipid II flippase MurJ, outward-facing conformation | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Lipid II flippase MurJ, ... | | Authors: | Kuk, A.C.Y, Lee, S.-Y. | | Deposit date: | 2018-12-11 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Visualizing conformation transitions of the Lipid II flippase MurJ.
Nat Commun, 10, 2019
|
|
6BW5
 
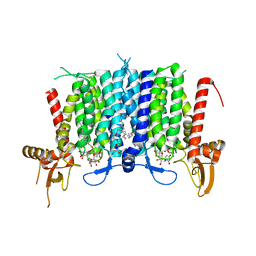 | | Human GPT (DPAGT1) in complex with tunicamycin | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Tunicamycin, UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase | | Authors: | Yoo, J, Kuk, A.C.Y, Mashalidis, E.H, Lee, S.-Y. | | Deposit date: | 2017-12-14 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | GlcNAc-1-P-transferase-tunicamycin complex structure reveals basis for inhibition of N-glycosylation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6BW6
 
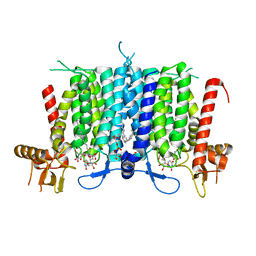 | | Human GPT (DPAGT1) H129 variant in complex with tunicamycin | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Tunicamycin, UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase | | Authors: | Yoo, J, Kuk, A.C.Y, Mashalidis, E.H, Lee, S.-Y. | | Deposit date: | 2017-12-14 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | GlcNAc-1-P-transferase-tunicamycin complex structure reveals basis for inhibition of N-glycosylation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
