2JZF
 
 | | NMR Conformer closest to the mean coordinates of the domain 513-651 of the SARS-CoV nonstructural protein nsp3 | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Chatterjee, A, Johnson, M.A, Serrano, P, Pedrini, B, Joseph, J, Saikatendu, K, Neuman, B, Stevens, R.C, Wilson, I.A, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-01-04 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold.
J.Virol., 83, 2009
|
|
2QBX
 
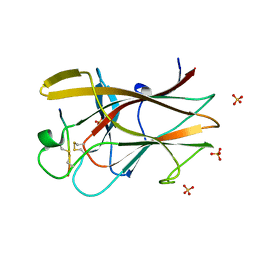 | | EphB2/SNEW Antagonistic Peptide Complex | | Descriptor: | Ephrin type-B receptor 2, SULFATE ION, antagonistic peptide | | Authors: | Chrencik, J.E, Brooun, A, Recht, M.I, Nicola, G, Pasquale, E.B, Kuhn, P, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2007-06-18 | | Release date: | 2007-11-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structure of the EphB2 receptor in complex with an antagonistic peptide reveals a novel mode of inhibition.
J.Biol.Chem., 282, 2007
|
|
2FYG
 
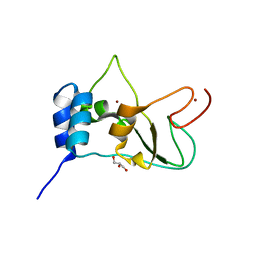 | | Crystal structure of NSP10 from Sars coronavirus | | Descriptor: | GLYCEROL, Replicase polyprotein 1ab, ZINC ION | | Authors: | Joseph, J.S, Saikatendu, K.S, Subramanian, V, Neuman, B.W, Brooun, A, Griffith, M, Moy, K, Yadav, M.K, Velasquez, J, Buchmeier, M.J, Stevens, R.C, Kuhn, P. | | Deposit date: | 2006-02-07 | | Release date: | 2006-08-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of nonstructural protein 10 from the severe acute respiratory syndrome coronavirus reveals a novel fold with two zinc-binding motifs.
J.Virol., 80, 2006
|
|
2RNK
 
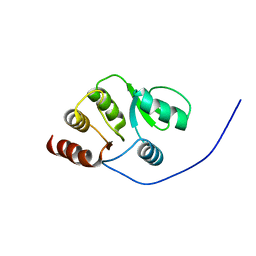 | | NMR structure of the domain 513-651 of the SARS-CoV nonstructural protein nsp3 | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Chatterjee, A, Johnson, M.A, Serrano, P, Pedrini, B, Joseph, J, Saikatendu, K, Neuman, B.W, Wilson, I.A, Stevens, R.C, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-01-11 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold.
J.Virol., 83, 2009
|
|
2X65
 
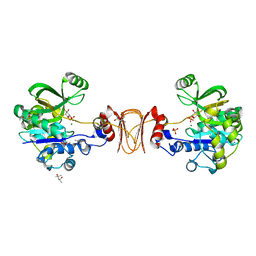 | | Crystal structure of T. maritima GDP-mannose pyrophosphorylase in complex with mannose-1-phosphate. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 1-O-phosphono-alpha-D-mannopyranose, ... | | Authors: | Pelissier, M.C, Lesley, S, Kuhn, P, Bourne, Y. | | Deposit date: | 2010-02-15 | | Release date: | 2010-06-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insights Into the Catalytic Mechanism of Bacterial Guanosine-Diphospho-D-Mannose Pyrophosphorylase and its Regulation by Divalent Ions.
J.Biol.Chem., 285, 2010
|
|
2HLE
 
 | |
2RH1
 
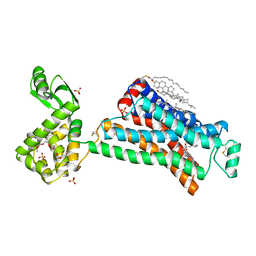 | | High resolution crystal structure of human B2-adrenergic G protein-coupled receptor. | | Descriptor: | (2S)-1-(9H-Carbazol-4-yloxy)-3-(isopropylamino)propan-2-ol, 1,4-BUTANEDIOL, ACETAMIDE, ... | | Authors: | Cherezov, V, Rosenbaum, D.M, Hanson, M.A, Rasmussen, S.G.F, Thian, F.S, Kobilka, T.S, Choi, H.J, Kuhn, P, Weis, W.I, Kobilka, B.K, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2007-10-05 | | Release date: | 2007-10-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor.
Science, 318, 2007
|
|
2X5S
 
 | |
2X5Z
 
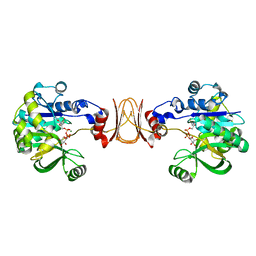 | | Crystal structure of T. maritima GDP-mannose pyrophosphorylase in complex with GDP-mannose. | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, MAGNESIUM ION, MANNOSE-1-PHOSPHATE GUANYLYLTRANSFERASE | | Authors: | Pelissier, M.C, Lesley, S, Kuhn, P, Bourne, Y. | | Deposit date: | 2010-02-12 | | Release date: | 2010-06-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insights Into the Catalytic Mechanism of Bacterial Guanosine-Diphospho-D-Mannose Pyrophosphorylase and its Regulation by Divalent Ions.
J.Biol.Chem., 285, 2010
|
|
1MX1
 
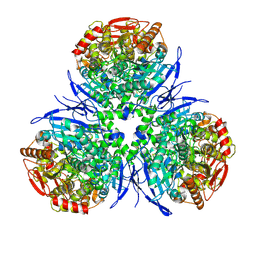 | | Crystal Structure of Human Liver Carboxylesterase in complex with tacrine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetyl-alpha-neuraminic acid, ... | | Authors: | Bencharit, S, Morton, C.L, Hyatt, J.L, Kuhn, P, Danks, M.K, Potter, P.M, Redinbo, M.R. | | Deposit date: | 2002-10-01 | | Release date: | 2003-04-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Human Carboxylesterase 1 Complexed with the Alzheimer's
Drug Tacrine: From Binding Promiscuity to Selective Inhibition
CHEM.BIOL., 10, 2003
|
|
2KAF
 
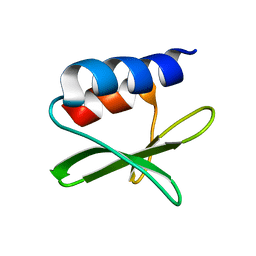 | | Solution structure of the SARS-unique domain-C from the nonstructural protein 3 (nsp3) of the severe acute respiratory syndrome coronavirus | | Descriptor: | Non-structural protein 3 | | Authors: | Johnson, M.A, Mohanty, B, Pedrini, B, Serrano, P, Chatterjee, A, Herrmann, T, Joseph, J, Saikatendu, K, Wilson, I.A, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-11-05 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | SARS coronavirus unique domain: three-domain molecular architecture in solution and RNA binding.
J.Mol.Biol., 400, 2010
|
|
2HSX
 
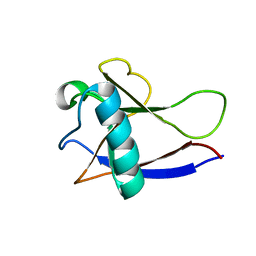 | | NMR Structure of the nonstructural protein 1 (nsp1) from the SARS coronavirus | | Descriptor: | Leader protein; p65 homolog; NSP1 (EC 3.4.22.-) | | Authors: | Almeida, M.S, Herrmann, T, Geralt, M, Johnson, M.A, Saikatendu, K, Joseph, J, Subramanian, R.C, Neuman, B.W, Buchmeier, M.J, Stevens, R.C, Kuhn, P, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2006-07-24 | | Release date: | 2007-02-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Novel beta-barrel fold in the nuclear magnetic resonance structure of the replicase nonstructural protein 1 from the severe acute respiratory syndrome coronavirus.
J.Virol., 81, 2007
|
|
2GDT
 
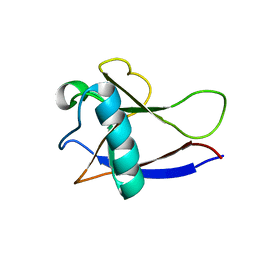 | | NMR Structure of the nonstructural protein 1 (nsp1) from the SARS coronavirus | | Descriptor: | Leader protein; p65 homolog; NSP1 (EC 3.4.22.-) | | Authors: | Almeida, M.S, Herrmann, T, Geralt, M, Johnson, M.A, Saikatendu, K, Joseph, J, Subramanian, R.C, Neuman, B.W, Buchmeier, M.J, Stevens, R.C, Kuhn, P, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2006-03-17 | | Release date: | 2007-02-06 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Novel beta-barrel fold in the nuclear magnetic resonance structure of the replicase nonstructural protein 1 from the severe acute respiratory syndrome coronavirus.
J.Virol., 81, 2007
|
|
5EFV
 
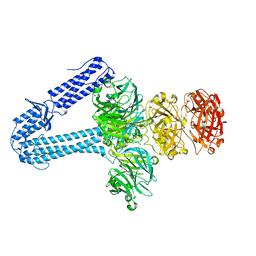 | | The host-recognition device of Staphylococcus aureus phage Phi11 | | Descriptor: | FE (III) ION, Phi ETA orf 56-like protein | | Authors: | Koc, C, Kuhner, P, Xia, G, Spinelli, S, Roussel, A, Cambillau, C, Stehle, T. | | Deposit date: | 2015-10-26 | | Release date: | 2016-05-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the host-recognition device of Staphylococcus aureus phage 11.
Sci Rep, 6, 2016
|
|
