1CFD
 
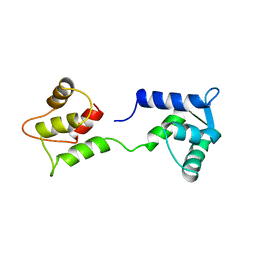 | | CALCIUM-FREE CALMODULIN | | Descriptor: | CALMODULIN | | Authors: | Kuboniwa, H, Tjandra, N, Grzesiek, S, Ren, H, Klee, C.B, Bax, A. | | Deposit date: | 1995-10-18 | | Release date: | 1995-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of calcium-free calmodulin.
Nat.Struct.Biol., 2, 1995
|
|
1CFC
 
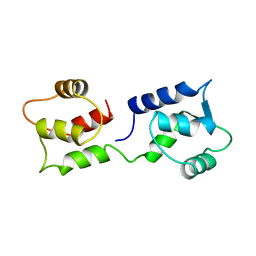 | | CALCIUM-FREE CALMODULIN | | Descriptor: | CALMODULIN | | Authors: | Kuboniwa, H, Tjandra, N, Grzesiek, S, Ren, H, Klee, C.B, Bax, A. | | Deposit date: | 1995-08-02 | | Release date: | 1995-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of calcium-free calmodulin.
Nat.Struct.Biol., 2, 1995
|
|
8ZYH
 
 | | Crystal structure of a cupin protein (tm1459, I49C-4py/H52A/H54A/C106D mutant) in copper (Cu) substituted form | | Descriptor: | 4-THIOPYRIDINE, COPPER (II) ION, Cupin type-2 domain-containing protein, ... | | Authors: | Morita, Y, Kubo, H, Matsumoto, R, Fujieda, N. | | Deposit date: | 2024-06-17 | | Release date: | 2024-09-04 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | A thiopyridine-bound mirror-image copper center in an artificial non-heme metalloenzyme.
J.Inorg.Biochem., 260, 2024
|
|
8ZYG
 
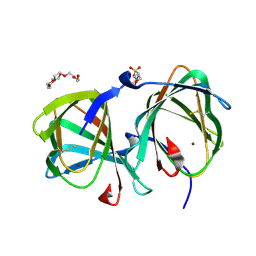 | | Crystal structure of a cupin protein (tm1459, I49C-4py/H52A/C106D mutant) in copper (Cu) substituted form | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, Cupin type-2 domain-containing protein, ... | | Authors: | Morita, Y, Kubo, H, Matsumoto, R, Fujieda, N. | | Deposit date: | 2024-06-17 | | Release date: | 2024-09-04 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | A thiopyridine-bound mirror-image copper center in an artificial non-heme metalloenzyme.
J.Inorg.Biochem., 260, 2024
|
|
7BUR
 
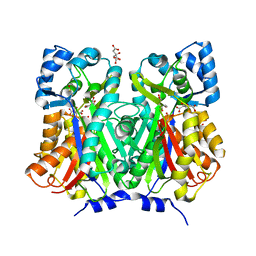 | | Chalcone synthase from Glycine max (L.) Merr (soybean) complexed with naringenin | | Descriptor: | CITRIC ACID, Chalcone synthase 1, NARINGENIN | | Authors: | Imaizumi, R, Mameda, R, Takeshita, K, Waki, T, Kubo, H, Sakai, N, Nakata, S, Takahashi, S, Kataoka, K, Yamamoto, M, Yamashita, S, Nakayama, T. | | Deposit date: | 2020-04-08 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of chalcone synthase, a key enzyme for isoflavonoid biosynthesis in soybean.
Proteins, 2020
|
|
7BUS
 
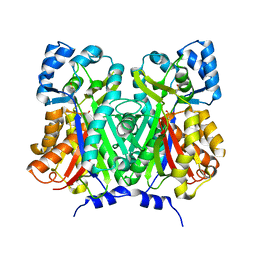 | | Chalcone synthase from Glycine max (L.) Merr (soybean) | | Descriptor: | Chalcone synthase | | Authors: | Imaizumi, R, Mameda, R, Takeshita, K, Waki, T, Kubo, H, Sakai, N, Nakata, S, Takahashi, S, Kataoka, K, Yamamoto, M, Yamashita, S, Nakayama, T. | | Deposit date: | 2020-04-08 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structure of chalcone synthase, a key enzyme for isoflavonoid biosynthesis in soybean.
Proteins, 2020
|
|
2RQY
 
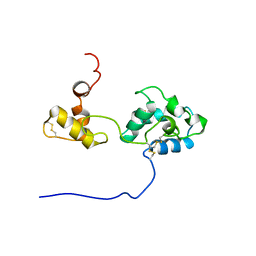 | | Solution structure and dynamics of mouse ARMET | | Descriptor: | Putative uncharacterized protein | | Authors: | Hoseki, J, Sasakawa, H, Yamaguchi, Y, Maeda, M, Kubota, H, Kato, K, Nagata, K. | | Deposit date: | 2010-01-26 | | Release date: | 2010-04-21 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of mouse ARMET.
Febs Lett., 584, 2010
|
|
6AIT
 
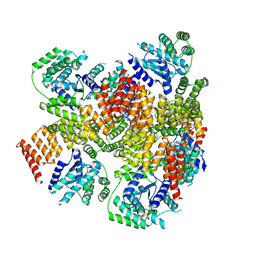 | | Crystal structure of E. coli BepA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-barrel assembly-enhancing protease, ZINC ION | | Authors: | Umar, M.S.M, Tanaka, Y, Kamikubo, H, Tsukazaki, T. | | Deposit date: | 2018-08-24 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Structural Basis for the Function of the beta-Barrel Assembly-Enhancing Protease BepA.
J. Mol. Biol., 431, 2019
|
|
5AUR
 
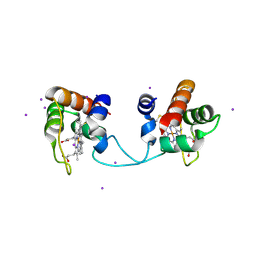 | | Hydrogenobacter thermophilus cytochrome c552 dimer formed by domain swapping at N-terminal region | | Descriptor: | Cytochrome c-552, HEME C, IODIDE ION | | Authors: | Ren, C, Nagao, S, Yamanaka, M, Kamikubo, H, Komori, H, Shomura, Y, Higuchi, Y, Hirota, S. | | Deposit date: | 2015-06-08 | | Release date: | 2015-10-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Oligomerization enhancement and two domain swapping mode detection for thermostable cytochrome c552via the elongation of the major hinge loop.
Mol Biosyst, 11, 2015
|
|
7VF8
 
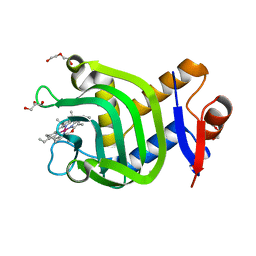 | | Crystal Structure of HasAp with Co-5-octaethyloxaporphyrinium cation | | Descriptor: | CITRIC ACID, Co-5-octaethyloxaporphyrinium cation, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Takiguchi, A, Sakakibara, E, Sugimoto, H, Shoji, O, Shinokubo, H. | | Deposit date: | 2021-09-10 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure of HasAp with Co-5-octaethyloxaporphyrinium cation
To Be Published
|
|
7VF7
 
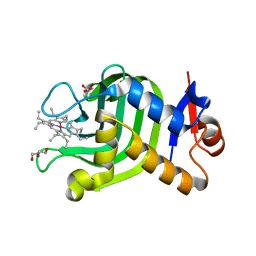 | | Crystal Structure of HasAp with Co-octaethylporphyrin | | Descriptor: | Co-octaethylporphyrin, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Takiguchi, A, Sakakibara, E, Sugimoto, H, Shoji, O, Shinokubo, H. | | Deposit date: | 2021-09-10 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure of HasAp with Co-octaethylporphyrin
To Be Published
|
|
1HNR
 
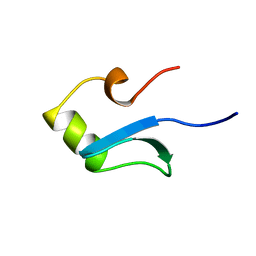 | | H-NS (DNA-BINDING DOMAIN) | | Descriptor: | H-NS | | Authors: | Shindo, H, Iwaki, T, Ieda, R, Kurumizaka, H, Ueguchi, C, Mizuno, T, Morikawa, S, Nakamura, H, Kuboniwa, H. | | Deposit date: | 1995-04-06 | | Release date: | 1995-07-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA binding domain of a nucleoid-associated protein, H-NS, from Escherichia coli.
FEBS Lett., 360, 1995
|
|
2IEZ
 
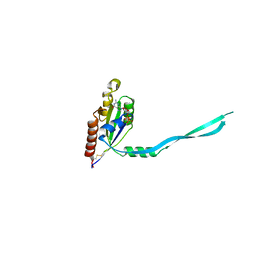 | | Crystal Structure of mouse Rab27b bound to GDP in monoclinic space group | | Descriptor: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, Ras-related protein Rab-27B | | Authors: | Chavas, L.M.G, Torii, S, Kamikubo, H, Kawasaki, M, Ihara, K, Kato, R, Kataoka, M, Izumi, T, Wakatsuki, S. | | Deposit date: | 2006-09-19 | | Release date: | 2007-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the small GTPase Rab27b shows an unexpected swapped dimer
Acta Crystallogr.,Sect.D, 63, 2007
|
|
1HNS
 
 | | H-NS (DNA-BINDING DOMAIN) | | Descriptor: | H-NS | | Authors: | Shindo, H, Iwaki, T, Ieda, R, Kurumizaka, H, Ueguchi, C, Mizuno, T, Morikawa, S, Nakamura, H, Kuboniwa, H. | | Deposit date: | 1995-04-06 | | Release date: | 1995-07-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA binding domain of a nucleoid-associated protein, H-NS, from Escherichia coli.
FEBS Lett., 360, 1995
|
|
2IF0
 
 | | Crystal Structure of mouse Rab27b bound to GDP in monoclinic space group | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-27B | | Authors: | Chavas, L.M.G, Torii, S, Kamikubo, H, Kawasaki, M, Ihara, K, Kato, R, Kataoka, M, Izumi, T, Wakatsuki, S. | | Deposit date: | 2006-09-19 | | Release date: | 2007-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the small GTPase Rab27b shows an unexpected swapped dimer
Acta Crystallogr.,Sect.D, 63, 2007
|
|
5H18
 
 | | Crystal structure of catalytic domain of UGGT (UDP-glucose-bound form) from Thermomyces dupontii | | Descriptor: | CALCIUM ION, GLYCEROL, UGGT, ... | | Authors: | Satoh, T, Zhu, T, Toshimori, T, Kamikubo, H, Uchihashi, T, Kato, K. | | Deposit date: | 2016-10-08 | | Release date: | 2017-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Visualisation of a flexible modular structure of the ER folding-sensor enzyme UGGT.
Sci Rep, 7, 2017
|
|
3X39
 
 | | Domain-swapped dimer of Pseudomonas aeruginosa cytochrome c551 | | Descriptor: | Cytochrome c-551, HEME C | | Authors: | Nagao, S, Ueda, M, Osuka, H, Komori, H, Kamikubo, H, Kataoka, M, Higuchi, Y, Hirota, S. | | Deposit date: | 2015-01-16 | | Release date: | 2015-04-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Domain-Swapped Dimer of Pseudomonas aeruginosa Cytochrome c551: Structural Insights into Domain Swapping of Cytochrome c Family Proteins
Plos One, 10, 2015
|
|
3VHX
 
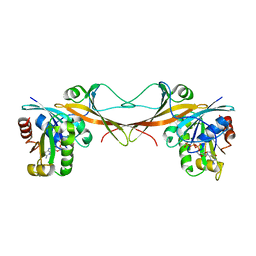 | | The crystal structure of Arf6-MKLP1 (Mitotic kinesin-like protein 1) complex | | Descriptor: | ADP-ribosylation factor 6, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Makyio, H, Takei, T, Ohgi, H, Takahashi, S, Takatsu, H, Ueda, T, Kanaho, Y, Xie, Y, Shin, H.W, Kamikubo, H, Kataoka, M, Kawasaki, M, Kato, R, Wakatsuki, S, Nakayama, K. | | Deposit date: | 2011-09-12 | | Release date: | 2012-05-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural basis for Arf6-MKLP1 complex formation on the Flemming body responsible for cytokinesis
Embo J., 31, 2012
|
|
2IEY
 
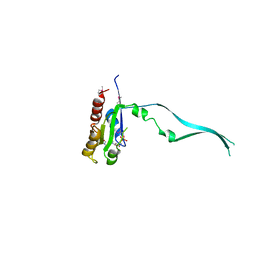 | | Crystal Structure of mouse Rab27b bound to GDP in hexagonal space group | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Ras-related protein Rab-27B | | Authors: | Chavas, L.M.G, Torii, S, Kamikubo, H, Kawasaki, M, Ihara, K, Kato, R, Kataoka, M, Izumi, T, Wakatsuki, S. | | Deposit date: | 2006-09-19 | | Release date: | 2007-05-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Structure of the small GTPase Rab27b shows an unexpected swapped dimer
Acta Crystallogr.,Sect.D, 63, 2007
|
|
3WUI
 
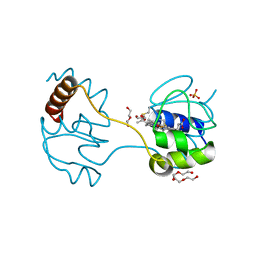 | | Dimeric horse cytochrome c formed by refolding from molten globule state | | Descriptor: | Cytochrome c, DI(HYDROXYETHYL)ETHER, HEME C, ... | | Authors: | Deshpande, M.S, Parui, P.P, Kamikubo, H, Yamanaka, M, Nagao, S, Komori, H, Kataoka, M, Higuchi, Y, Hirota, S. | | Deposit date: | 2014-04-25 | | Release date: | 2014-07-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Formation of domain-swapped oligomer of cytochrome C from its molten globule state oligomer.
Biochemistry, 53, 2014
|
|
4BBT
 
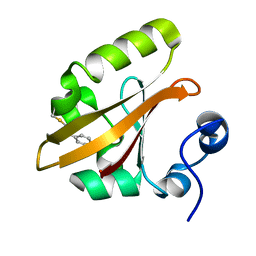 | | The PR1 Photocycle Intermediate of Photoactive Yellow Protein | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Schotte, F, Cho, H.S, Kaila, V.R.I, Kamikubo, H, Dashdorj, N, Henry, E.R, Graber, T.J, Henning, R, Wulff, M, Hummer, G, Kataoka, M, Anfinrud, P.A. | | Deposit date: | 2012-09-27 | | Release date: | 2012-11-14 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Watching a Signaling Protein Function in Real Time Via 100-Ps Time-Resolved Laue Crystallography.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4B9O
 
 | | The PR0 Photocycle Intermediate of Photoactive Yellow Protein | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Schotte, F, Cho, H.S, Kaila, V.R.I, Kamikubo, H, Dashdorj, N, Henry, E.R, Graber, T.J, Henning, R, Wulff, M, Hummer, G, Kataoka, M, Anfinrud, P.A. | | Deposit date: | 2012-09-06 | | Release date: | 2012-11-14 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Watching a Signaling Protein Function in Real Time Via 100-Ps Time-Resolved Laue Crystallography
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4BBU
 
 | | The PR2 Photocycle Intermediate of Photoactive Yellow Protein | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Schotte, F, Cho, H.S, Kaila, V.R.I, Kamikubo, H, Dashdorj, N, Henry, E.R, Graber, T.J, Henning, R, Wulff, M, Hummer, G, Kataoka, M, Anfinrud, P.A. | | Deposit date: | 2012-09-27 | | Release date: | 2012-11-14 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Watching a Signaling Protein Function in Real Time Via 100-Ps Time-Resolved Laue Crystallography.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4BBV
 
 | | The PB0 Photocycle Intermediate of Photoactive Yellow Protein | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Schotte, F, Cho, H.S, Kaila, V.R.I, Kamikubo, H, Dashdorj, N, Henry, E.R, Graber, T.J, Henning, R, Wulff, M, Hummer, G, Kataoka, M, Anfinrud, P.A. | | Deposit date: | 2012-09-28 | | Release date: | 2012-11-14 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Watching a Signaling Protein Function in Real Time Via 100-Ps Time-Resolved Laue Crystallography.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3WC8
 
 | | Dimeric horse cytochrome c obtained by refolding with desalting method | | Descriptor: | Cytochrome c, DI(HYDROXYETHYL)ETHER, HEME C, ... | | Authors: | Parui, P.P, Deshpande, M.S, Nagao, S, Kamikubo, H, Komori, H, Higuchi, Y, Kataoka, M, Hirota, S. | | Deposit date: | 2013-05-25 | | Release date: | 2013-12-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Formation of Oligomeric Cytochrome c during Folding by Intermolecular Hydrophobic Interaction between N- and C-Terminal alpha-Helices
Biochemistry, 52, 2013
|
|
