1SHK
 
 | |
2SHK
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF SHIKIMATE KINASE FROM ERWINIA CHRYSANTHEMI COMPLEXED WITH ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SHIKIMATE KINASE | | Authors: | Krell, T, Coggins, J.R, Lapthorn, A.J. | | Deposit date: | 1997-10-27 | | Release date: | 1998-11-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallization and preliminary X-ray crystallographic analysis of shikimate kinase from Erwinia chrysanthemi.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
8BMV
 
 | | Ligand binding domain of the P. Putida receptor McpH in complex with Uric acid | | Descriptor: | Methyl-accepting chemotaxis protein McpH, URIC ACID | | Authors: | Gavira, J.A, Krell, T, Fernandez, M, Martinez-Rodriguez, S. | | Deposit date: | 2022-11-11 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Ubiquitous purine sensor modulates diverse signal transduction pathways in bacteria.
Nat Commun, 15, 2024
|
|
6F9G
 
 | | Ligand binding domain of P. putida KT2440 polyamine chemorecpetors McpU in complex putrescine. | | Descriptor: | 1,4-DIAMINOBUTANE, ACETATE ION, GLYCEROL, ... | | Authors: | Gavira, J.A, Conejero-Muriel, M.T, Ortega, A, Martin-Mora, D, Corral-Lugo, A, Morel, B, Krell, T. | | Deposit date: | 2017-12-14 | | Release date: | 2018-03-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.388 Å) | | Cite: | Structural Basis for Polyamine Binding at the dCACHE Domain of the McpU Chemoreceptor from Pseudomonas putida.
J. Mol. Biol., 430, 2018
|
|
6YMZ
 
 | | Structure of the CheB methylsterase from P. atrosepticum SCRI1043 | | Descriptor: | ACETATE ION, GLYCEROL, Protein-glutamate methylesterase/protein-glutamine glutaminase, ... | | Authors: | Gavira, J.A, Krell, T, Velando-Soriano, F, Matilla, M.A. | | Deposit date: | 2020-04-10 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evidence for Pentapeptide-Dependent and Independent CheB Methylesterases.
Int J Mol Sci, 21, 2020
|
|
5LTV
 
 | | LIGAND BINDING DOMAIN OF PSEUDOMONAS AERUGINOSA PAO1 AMINO ACID CHEMORECPETOR PCTC IN COMPLEX WITH GABA | | Descriptor: | ACETATE ION, Chemotactic transducer PctC, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Gavira, J.A, Rico-Jimenez, M, Conejero-Muriel, M, Krell, T. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | How Bacterial Chemoreceptors Evolve Novel Ligand Specificities
Mbio, 2020
|
|
1E6C
 
 | | K15M MUTANT OF SHIKIMATE KINASE FROM ERWINIA CHRYSANTHEMI | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | Maclean, J, Krell, T, Coggins, J.R, Lapthorn, A.J. | | Deposit date: | 2000-08-10 | | Release date: | 2001-06-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical and X-Ray Crystallographic Studies on Shikimate Kinase: The Important Structural Role of the P-Loop Lysine
Protein Sci., 10, 2001
|
|
1D0I
 
 | | CRYSTAL STRUCTURE OF TYPE II DEHYDROQUINASE FROM STREPTOMYCES COELICOLOR COMPLEXED WITH PHOSPHATE IONS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHOSPHATE ION, TYPE II 3-DEHYDROQUINATE HYDRATASE | | Authors: | Roszak, A.W, Krell, T, Hunter, I.S, Coggins, J.R, Lapthorn, A.J. | | Deposit date: | 1999-09-10 | | Release date: | 2000-09-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure and mechanism of the type II dehydroquinase from Streptomyces coelicolor.
Structure, 10, 2002
|
|
7PSG
 
 | | Structure of the ligand binding domain of the PacA (ECA2226) chemoreceptor of Pectobacterium atrosepticum SCRI1043 in complex with betaine. | | Descriptor: | GLYCEROL, Methyl-accepting chemotaxis protein, TRIMETHYL GLYCINE | | Authors: | Gavira, J.A, Matilla, M.A, Velando, F, Krell, T. | | Deposit date: | 2021-09-23 | | Release date: | 2022-05-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Chemotaxis of the Human Pathogen Pseudomonas aeruginosa to the Neurotransmitter Acetylcholine.
Mbio, 13, 2022
|
|
5T65
 
 | | LIGAND BINDING DOMAIN OF PSEUDOMONAS AERUGINOSA PAO1 AMINO ACID CHEMORECEPTOR PCTA IN COMPLEX WITH L-ILE | | Descriptor: | ACETATE ION, ISOLEUCINE, Methyl-accepting chemotaxis protein PctA, ... | | Authors: | Gavira, J.A, Rico-Jimenez, M, Ortega, A, Conejero-Muriel, M, Zhulin, I, Krell, T. | | Deposit date: | 2016-09-01 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | How Bacterial Chemoreceptors Evolve Novel Ligand Specificities
Mbio, 2020
|
|
5LTO
 
 | | Ligand binding domain of Pseudomonas aeruginosa PAO1 amino acid chemoreceptors PctB in complex with L-Gln | | Descriptor: | GLUTAMINE, GLYCEROL, Methyl-accepting chemotaxis protein PctB, ... | | Authors: | Gavira, J.A, Rico-Jimenez, M, Conejero-Muriel, M, Krell, T. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.459 Å) | | Cite: | How Bacterial Chemoreceptors Evolve Novel Ligand Specificities
Mbio, 2020
|
|
5LT9
 
 | | Ligand binding domain of Pseudomonas aeruginosa PAO1 amino acid chemoreceptors PctB in complex with L-Arg | | Descriptor: | ARGININE, GLYCEROL, Methyl-accepting chemotaxis protein PctB, ... | | Authors: | Gavira, J.A, Rico-Jimenez, M, Ortega, A, Conejero-Muriel, M, Zhulin, I, Krell, T. | | Deposit date: | 2016-09-06 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | How Bacterial Chemoreceptors Evolve Novel Ligand Specificities
Mbio, 2020
|
|
5T7M
 
 | | LIGAND BINDING DOMAIN OF PSEUDOMONAS AERUGINOSA PAO1 AMINO ACID CHEMORECEPTOR PCTA IN COMPLEX WITH L-TRP | | Descriptor: | ACETATE ION, Chemotaxis protein, SODIUM ION, ... | | Authors: | Gavira, J.A, Rico-Jimenez, M, Ortega, A, Conejero-Muriel, M, Zhulin, I, Krell, T. | | Deposit date: | 2016-09-05 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | How Bacterial Chemoreceptors Evolve Novel Ligand Specificities
Mbio, 2020
|
|
7QEK
 
 | | Structure of the ligand binding domain of the antibiotic biosynthesis regulator AdmX from the rhizobacterium Serratia plymuthica A153 bound to the auxin indole-3-piruvic acid (IPA). | | Descriptor: | 3-(1H-INDOL-3-YL)-2-OXOPROPANOIC ACID, MAGNESIUM ION, regulator AdmX | | Authors: | Gavira, J.A, Rico-Jimenez, M, Castellvi, A, Krell, T, Matilla, M.A. | | Deposit date: | 2021-12-03 | | Release date: | 2022-12-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Emergence of an Auxin Sensing Domain in Plant-Associated Bacteria.
Mbio, 14, 2023
|
|
7QEJ
 
 | | Structure of the ligand binding domain of the antibiotic biosynthesis regulator AdmX from the rhizobacterium Serratia plymuthica A153 bound to the auxin indole-3-acetic acid (IAA). | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, MAGNESIUM ION, TRANSCRIPTIONAL REGULATOR AdmX | | Authors: | Gavira, J.A, Rico-Jimenez, M, Castellvi, A, Krell, T, Matilla, M.A. | | Deposit date: | 2021-12-03 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Emergence of an Auxin Sensing Domain in Plant-Associated Bacteria.
Mbio, 14, 2023
|
|
6FU4
 
 | | Ligand binding domain (LBD) of the p. aeruginosa histamine receptor TlpQ | | Descriptor: | ACETATE ION, GLYCEROL, HISTAMINE, ... | | Authors: | Gavira, J.A, Krell, T, Conejero-Muriel, M, Corral-Lugo, A, Matilla, M.A, Silva Jimenez, H, Mesa Torres, N, Martin-Mora, D. | | Deposit date: | 2018-02-26 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | High-Affinity Chemotaxis to Histamine Mediated by the TlpQ Chemoreceptor of the Human Pathogen Pseudomonas aeruginosa.
MBio, 9, 2018
|
|
6GCV
 
 | | Ligand binding domain (LBD) of the p. aeruginosa nitrate receptor McpN | | Descriptor: | ACETATE ION, Chemotaxis transducer, NITRATE ION, ... | | Authors: | Gavira, J.A, Krell, T, Martin-Mora, D, Ortega, A. | | Deposit date: | 2018-04-19 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Molecular Mechanism of Nitrate Chemotaxis via Direct Ligand Binding to the PilJ Domain of McpN.
MBio, 10, 2019
|
|
7PRR
 
 | | Structure of the ligand binding domain of the PctD (PA4633) chemoreceptor of Pseudomonas aeruginosa PAO1 in complex with acetylcholine | | Descriptor: | ACETYLCHOLINE, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gavira, J.A, Matilla, M.A, Martin-Mora, D, Krell, T. | | Deposit date: | 2021-09-22 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Chemotaxis of the Human Pathogen Pseudomonas aeruginosa to the Neurotransmitter Acetylcholine.
Mbio, 13, 2022
|
|
7PRQ
 
 | | Structure of the ligand binding domain of the PctD (PA4633) chemoreceptor of Pseudomonas aeruginosa PAO1 in complex with choline. | | Descriptor: | 1,2-ETHANEDIOL, CHOLINE ION, GLYCEROL, ... | | Authors: | Gavira, J.A, Matilla, M.A, Martin-Mora, D, Krell, T. | | Deposit date: | 2021-09-22 | | Release date: | 2022-05-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Chemotaxis of the Human Pathogen Pseudomonas aeruginosa to the Neurotransmitter Acetylcholine.
Mbio, 13, 2022
|
|
2YFB
 
 | | X-ray structure of McpS ligand binding domain in complex with succinate | | Descriptor: | ACETATE ION, METHYL-ACCEPTING CHEMOTAXIS TRANSDUCER, SUCCINIC ACID, ... | | Authors: | Gavira, J.A, Pineda-Molina, E, Krell, T. | | Deposit date: | 2011-04-05 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evidence for Chemoreceptors with Bimodular Ligand-Binding Regions Harboring Two Signal-Binding Sites.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2YFA
 
 | | X-ray structure of McpS ligand binding domain in complex with malate | | Descriptor: | (2S)-2-hydroxybutanedioic acid, ACETATE ION, METHYL-ACCEPTING CHEMOTAXIS TRANSDUCER, ... | | Authors: | Pineda-Molina, E, Gavira, J.A, Krell, T. | | Deposit date: | 2011-04-05 | | Release date: | 2012-04-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evidence for Chemoreceptors with Bimodular Ligand-Binding Regions Harboring Two Signal-Binding Sites.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5LTX
 
 | | LIGAND BINDING DOMAIN OF PSEUDOMONAS AERUGINOSA PAO1 AMINO ACID CHEMORECEPTOR PCTA IN COMPLEX WITH L-MET | | Descriptor: | ACETATE ION, Chemotaxis protein, FORMIC ACID, ... | | Authors: | Gavira, J.A, Rico-Gimenez, M, Ortega, A, Conejero-Muriel, M, Zhulin, I, Krell, T. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | How Bacterial Chemoreceptors Evolve Novel Ligand Specificities
Mbio, 2020
|
|
3O75
 
 | | Crystal structure of Cra transcriptional dual regulator from Pseudomonas putida in complex with fructose 1-phosphate' | | Descriptor: | 1-O-phosphono-beta-D-fructofuranose, Fructose transport system repressor FruR | | Authors: | Chavarria, M, Santiago, C, Platero, R, Krell, T, Casasnovas, J.M, de Lorenzo, V. | | Deposit date: | 2010-07-30 | | Release date: | 2011-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fructose 1-phosphate is the preferred effector of the metabolic regulator Cra of Pseudomonas putida
J.Biol.Chem., 286, 2011
|
|
3O74
 
 | | Crystal structure of Cra transcriptional dual regulator from Pseudomonas putida | | Descriptor: | Fructose transport system repressor FruR, GLYCEROL | | Authors: | Chavarria, M, Santiago, C, Platero, R, Krell, T, Casasnovas, J.M, de Lorenzo, V. | | Deposit date: | 2010-07-30 | | Release date: | 2011-01-12 | | Last modified: | 2011-12-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fructose 1-phosphate is the preferred effector of the metabolic regulator Cra of Pseudomonas putida
J.Biol.Chem., 286, 2011
|
|
2XRO
 
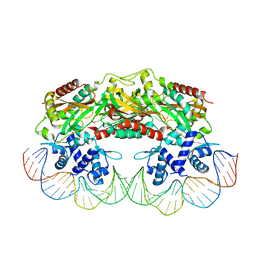 | | Crystal structure of TtgV in complex with its DNA operator | | Descriptor: | HTH-TYPE TRANSCRIPTIONAL REGULATOR TTGV, OSMIUM ION, TTGV OPERATOR DNA | | Authors: | Lu, D, Fillet, S, Meng, C, Alguel, Y, Kloppsteck, P, Bergeron, J, Krell, T, Gallegos, M.-T, Ramos, J, Zhang, X. | | Deposit date: | 2010-09-17 | | Release date: | 2010-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal Structure of Ttgv in Complex with its DNA Operator Reveals a General Model for Cooperative DNA Binding of Tetrameric Gene Regulators.
Genes Dev., 24, 2010
|
|
