1QJ4
 
 | |
7YAS
 
 | | HYDROXYNITRILE LYASE, LOW TEMPERATURE NATIVE STRUCTURE | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, PROTEIN (HYDROXYNITRILE LYASE), ... | | Authors: | Zuegg, J, Wagner, U.G, Gugganig, M, Kratky, C. | | Deposit date: | 1999-03-15 | | Release date: | 1999-10-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Three-dimensional structures of enzyme-substrate complexes of the hydroxynitrile lyase from Hevea brasiliensis.
Protein Sci., 8, 1999
|
|
5YAS
 
 | | HYDROXYNITRILE LYASE COMPLEXED WITH HEXAFLUOROACETONE | | Descriptor: | 1,1,1,3,3,3-HEXAFLUOROPROPANEDIOL, PROTEIN (HYDROXYNITRILE LYASE), SULFATE ION | | Authors: | Zuegg, J, Wagner, U.G, Gugganig, M, Kratky, C. | | Deposit date: | 1999-03-15 | | Release date: | 1999-10-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structures of enzyme-substrate complexes of the hydroxynitrile lyase from Hevea brasiliensis.
Protein Sci., 8, 1999
|
|
6YAS
 
 | | HYDROXYNITRILE LYASE FROM HEVEA BRASILIENSIS, ROOM TEMPERATURE STRUCTURE | | Descriptor: | PROTEIN (HYDROXYNITRILE LYASE), SULFATE ION | | Authors: | Zuegg, J, Wagner, U.G, Gugganig, M, Kratky, C. | | Deposit date: | 1999-03-15 | | Release date: | 1999-10-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structures of enzyme-substrate complexes of the hydroxynitrile lyase from Hevea brasiliensis.
Protein Sci., 8, 1999
|
|
4YAS
 
 | | HYDROXYNITRILE LYASE COMPLEXED WITH CHLORALHYDRATE | | Descriptor: | PROTEIN (HYDROXYNITRILE LYASE), SULFATE ION, TRI-CHLORO-ACETALDEHYDE | | Authors: | Zuegg, J, Wagner, U.G, Gugganig, M, Kratky, C. | | Deposit date: | 1999-03-15 | | Release date: | 1999-10-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structures of enzyme-substrate complexes of the hydroxynitrile lyase from Hevea brasiliensis.
Protein Sci., 8, 1999
|
|
3YAS
 
 | | HYDROXYNITRILE LYASE COMPLEXED WITH ACETONE | | Descriptor: | ACETONE, PROTEIN (HYDROXYNITRILE LYASE), SULFATE ION | | Authors: | Zuegg, J, Wagner, U.G, Gugganig, M, Kratky, C. | | Deposit date: | 1999-03-15 | | Release date: | 1999-10-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Three-dimensional structures of enzyme-substrate complexes of the hydroxynitrile lyase from Hevea brasiliensis.
Protein Sci., 8, 1999
|
|
1I9C
 
 | |
2YAS
 
 | |
3GDN
 
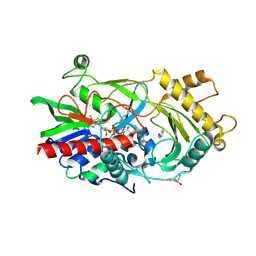 | | Almond hydroxynitrile lyase in complex with benzaldehyde | | Descriptor: | (2R)-hydroxy(phenyl)ethanenitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dreveny, I, Gruber, K, Kratky, C. | | Deposit date: | 2009-02-24 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Substrate binding in the FAD-dependent hydroxynitrile lyase from almond provides insight into the mechanism of cyanohydrin formation and explains the absence of dehydrogenation activity.
Biochemistry, 48, 2009
|
|
1SCI
 
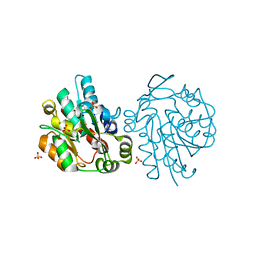 | | K236L mutant of hydroxynitrile lyase from Hevea brasiliensis | | Descriptor: | (S)-acetone-cyanohydrin lyase, SULFATE ION | | Authors: | Gruber, K, Gartler, G, Krammer, B, Schwab, H, Kratky, C. | | Deposit date: | 2004-02-12 | | Release date: | 2004-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Reaction mechanism of hydroxynitrile lyases of the alpha/beta-hydrolase superfamily: the three-dimensional structure of the transient enzyme-substrate complex certifies the crucial role of LYS236
J.Biol.Chem., 279, 2004
|
|
1SCK
 
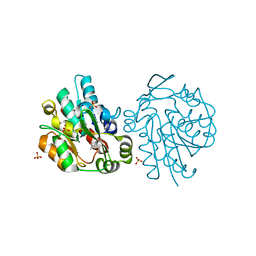 | | K236L mutant of hydroxynitrile lyase from Hevea brasiliensis in complex with acetone | | Descriptor: | (S)-acetone-cyanohydrin lyase, ACETONE, SULFATE ION | | Authors: | Gruber, K, Gartler, G, Krammer, B, Schwab, H, Kratky, C. | | Deposit date: | 2004-02-12 | | Release date: | 2004-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Reaction mechanism of hydroxynitrile lyases of the alpha/beta-hydrolase superfamily: the three-dimensional structure of the transient enzyme-substrate complex certifies the crucial role of LYS236
J.Biol.Chem., 279, 2004
|
|
1SCQ
 
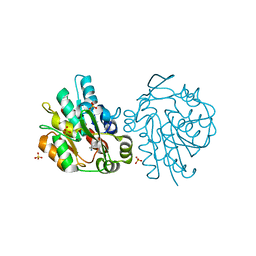 | | K236L mutant of hydroxynitrile lyase from Hevea brasiliensis in complex with acetonecyanohydrin | | Descriptor: | (S)-acetone-cyanohydrin lyase, 2-HYDROXY-2-METHYLPROPANENITRILE, SULFATE ION | | Authors: | Gruber, K, Gartler, G, Krammer, B, Schwab, H, Kratky, C. | | Deposit date: | 2004-02-12 | | Release date: | 2004-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Reaction mechanism of hydroxynitrile lyases of the alpha/beta-hydrolase superfamily: the three-dimensional structure of the transient enzyme-substrate complex certifies the crucial role of LYS236
J.Biol.Chem., 279, 2004
|
|
1YB6
 
 | |
1YB7
 
 | | Hydroxynitrile lyase from hevea brasiliensis in complex with 2,3-dimethyl-2-hydroxy-butyronitrile | | Descriptor: | (S)-2-HYDROXY-2,3-DIMETHYLBUTANENITRILE, (S)-acetone-cyanohydrin lyase, SULFATE ION | | Authors: | Gruber, K, Gartler, G, Kratky, C. | | Deposit date: | 2004-12-20 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural determinants of the enantioselectivity of the hydroxynitrile lyase from Hevea brasiliensis
J.Biotechnol., 129, 2007
|
|
1SC9
 
 | | Hydroxynitrile Lyase from Hevea brasiliensis in complex with the natural substrate acetone cyanohydrin | | Descriptor: | (S)-acetone-cyanohydrin lyase, 2-HYDROXY-2-METHYLPROPANENITRILE, SULFATE ION | | Authors: | Gruber, K, Gartler, G, Krammer, B, Schwab, H, Kratky, C. | | Deposit date: | 2004-02-12 | | Release date: | 2004-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reaction mechanism of hydroxynitrile lyases of the alpha/beta-hydrolase superfamily: the three-dimensional structure of the transient enzyme-substrate complex certifies the crucial role of LYS236
J.Biol.Chem., 279, 2004
|
|
1JU2
 
 | | Crystal structure of the hydroxynitrile lyase from almond | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dreveny, I, Gruber, K, Glieder, A, Thompson, A, Kratky, C. | | Deposit date: | 2001-08-23 | | Release date: | 2002-09-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The hydroxynitrile lyase from almond: a lyase that looks like an oxidoreductase.
Structure, 9, 2001
|
|
3GDP
 
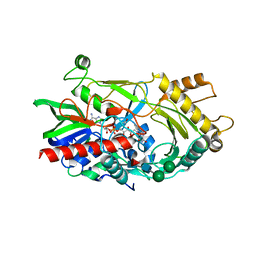 | | Hydroxynitrile lyase from almond, monoclinic crystal form | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dreveny, I, Gruber, K, Kratky, C. | | Deposit date: | 2009-02-24 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Substrate binding in the FAD-dependent hydroxynitrile lyase from almond provides insight into the mechanism of cyanohydrin formation and explains the absence of dehydrogenation activity.
Biochemistry, 48, 2009
|
|
1YAS
 
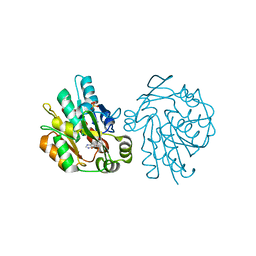 | | HYDROXYNITRILE LYASE COMPLEXED WITH HISTIDINE | | Descriptor: | HISTIDINE, HYDROXYNITRILE LYASE, SULFATE ION | | Authors: | Wagner, U.G, Kratky, C. | | Deposit date: | 1996-05-15 | | Release date: | 1997-06-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of cyanogenesis: the crystal structure of hydroxynitrile lyase from Hevea brasiliensis.
Structure, 4, 1996
|
|
3BAL
 
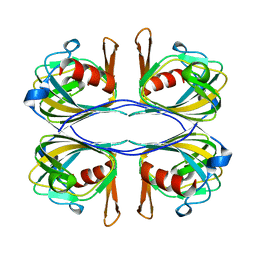 | | Crystal Structure of an Acetylacetone Dioxygenase from Acinetobacter johnsonii | | Descriptor: | Acetylacetone-cleaving enzyme, ZINC ION | | Authors: | Stranzl, G.R, Wagner, U.G, Straganz, G, Steiner, W, Kratky, C. | | Deposit date: | 2007-11-08 | | Release date: | 2007-11-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of an Acetylacetone Dioxygenase from Acinetobacter johnsonii
To be Published
|
|
6H9F
 
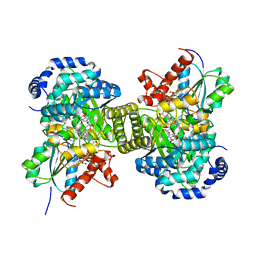 | | Structure of glutamate mutase reconstituted with bishomo-coenzyme B12 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-propyl-oxolane-3,4-diol, COBALAMIN, D(-)-TARTARIC ACID, ... | | Authors: | Gruber, K, Csitkovits, V, Kratky, C. | | Deposit date: | 2018-08-03 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Demystification of Radical Catalysis by a Coenzyme B 12 Dependent Enzyme-Crystallographic Study of Glutamate Mutase with Cofactor Homologues.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
6H9E
 
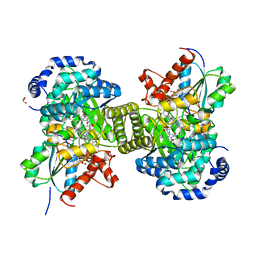 | | Structure of glutamate mutase reconstituted with homo-coenzyme B12 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-ethyl-oxolane-3,4-diol, COBALAMIN, D(-)-TARTARIC ACID, ... | | Authors: | Gruber, K, Csitkovits, V, Kratky, C. | | Deposit date: | 2018-08-03 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure-Based Demystification of Radical Catalysis by a Coenzyme B 12 Dependent Enzyme-Crystallographic Study of Glutamate Mutase with Cofactor Homologues.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
1CI8
 
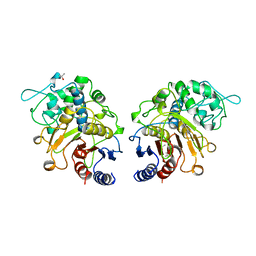 | | ESTERASE ESTB FROM BURKHOLDERIA GLADIOLI: AN ESTERASE WITH (BETA)-LACTAMASE FOLD. | | Descriptor: | ISOPROPYL ALCOHOL, PROTEIN (CARBOXYLESTERASE) | | Authors: | Wagner, U.G, Petersen, E.I, Schwab, H, Kratky, C. | | Deposit date: | 1999-04-08 | | Release date: | 2001-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | EstB from Burkholderia gladioli: a novel esterase with a beta-lactamase fold reveals steric factors to discriminate between esterolytic and beta-lactam cleaving activity
Protein Sci., 11, 2002
|
|
1CI9
 
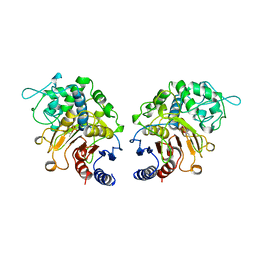 | | DFP-INHIBITED ESTERASE ESTB FROM BURKHOLDERIA GLADIOLI | | Descriptor: | DIISOPROPYL PHOSPHONATE, PROTEIN (CARBOXYLESTERASE) | | Authors: | Wagner, U.G, Petersen, E.I, Schwab, H, Kratky, C. | | Deposit date: | 1999-04-08 | | Release date: | 2001-12-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | EstB from Burkholderia gladioli: a novel esterase with a beta-lactamase fold reveals steric factors to discriminate between esterolytic and beta-lactam cleaving activity
Protein Sci., 11, 2002
|
|
1FR3
 
 | |
1YNA
 
 | | ENDO-1,4-BETA-XYLANASE, ROOM TEMPERATURE, PH 4.0 | | Descriptor: | ENDO-1,4-BETA-XYLANASE | | Authors: | Gruber, K, Kratky, C. | | Deposit date: | 1996-08-22 | | Release date: | 1997-02-12 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Thermophilic xylanase from Thermomyces lanuginosus: high-resolution X-ray structure and modeling studies.
Biochemistry, 37, 1998
|
|
