5TDA
 
 | |
5BTZ
 
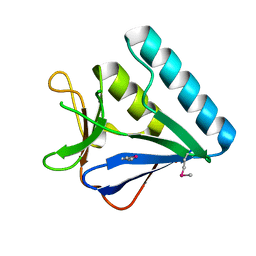 | | Structure of the middle domain of lpg1496 from Legionella pneumophila in P212121 space group | | Descriptor: | lpg1496 | | Authors: | Wong, K, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2015-06-03 | | Release date: | 2015-08-26 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the Legionella Effector, lpg1496, Suggests a Role in Nucleotide Metabolism.
J.Biol.Chem., 290, 2015
|
|
5BTX
 
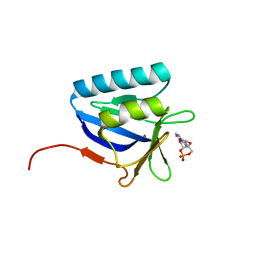 | | Structure of the N-terminal domain of lpg1496 from Legionella pneumophila in complex with nucleotide | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, lpg1496 | | Authors: | Wong, K, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2015-06-03 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the Legionella Effector, lpg1496, Suggests a Role in Nucleotide Metabolism.
J.Biol.Chem., 290, 2015
|
|
5BU0
 
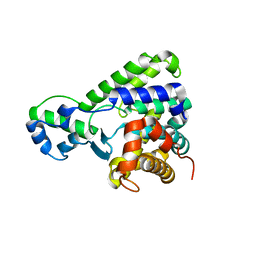 | | Structure of the C-terminal domain of lpg1496 from Legionella pneumophila | | Descriptor: | lpg1496 | | Authors: | Wong, K, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2015-06-03 | | Release date: | 2015-08-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of the Legionella Effector, lpg1496, Suggests a Role in Nucleotide Metabolism.
J.Biol.Chem., 290, 2015
|
|
5BTW
 
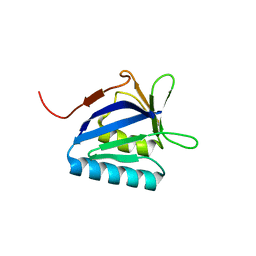 | | Structure of the N-terminal domain of lpg1496 from Legionella pneumophila | | Descriptor: | Uncharacterized protein | | Authors: | Wong, K, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2015-06-03 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of the Legionella Effector, lpg1496, Suggests a Role in Nucleotide Metabolism.
J.Biol.Chem., 290, 2015
|
|
5BU1
 
 | | Structure of the truncated C-terminal domain of lpg1496 from Legionella pneumophila | | Descriptor: | LPG1496, MALONATE ION | | Authors: | Wong, K, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2015-06-03 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the Legionella Effector, lpg1496, Suggests a Role in Nucleotide Metabolism.
J.Biol.Chem., 290, 2015
|
|
1JDQ
 
 | | Solution Structure of TM006 Protein from Thermotoga maritima | | Descriptor: | HYPOTHETICAL PROTEIN TM0983 | | Authors: | Denisov, A.Y, Finak, G, Yee, A, Kozlov, G, Gehring, K, Arrowsmith, C.H. | | Deposit date: | 2001-06-14 | | Release date: | 2002-02-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An NMR approach to structural proteomics.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1JE3
 
 | | Solution Structure of EC005 from Escherichia coli | | Descriptor: | HYPOTHETICAL 8.6 KDA PROTEIN IN AMYA-FLIE INTERGENIC REGION | | Authors: | Yee, A, Gutierrez, P, Kozlov, G, Denisov, A, Gehring, K, Arrowsmith, C. | | Deposit date: | 2001-06-15 | | Release date: | 2002-03-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An NMR approach to structural proteomics.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
4GWR
 
 | |
4EF0
 
 | |
3NY2
 
 | | Structure of the ubr-box of UBR2 ubiquitin ligase | | Descriptor: | E3 ubiquitin-protein ligase UBR2, ZINC ION | | Authors: | Matta-Camacho, E, Kozlov, G, Li, F, Gehring, K. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural basis of substrate recognition and specificity in the N-end rule pathway.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NY1
 
 | | Structure of the ubr-box of the UBR1 ubiquitin ligase | | Descriptor: | E3 ubiquitin-protein ligase UBR1, ZINC ION | | Authors: | Matta-Camacho, E, Kozlov, G, Li, F, Gehring, K. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.085 Å) | | Cite: | Structural basis of substrate recognition and specificity in the N-end rule pathway.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NY3
 
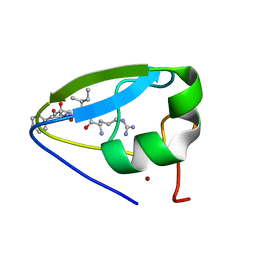 | | Structure of the ubr-box of UBR2 in complex with N-degron | | Descriptor: | E3 ubiquitin-protein ligase UBR2, N-degron, ZINC ION | | Authors: | Matta-Camacho, E, Kozlov, G, Li, F, Gehring, K. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of substrate recognition and specificity in the N-end rule pathway.
Nat.Struct.Mol.Biol., 17, 2010
|
|
5TDD
 
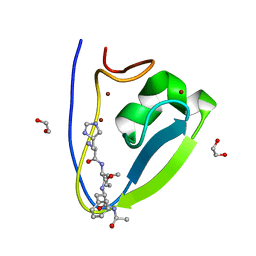 | | Human UBR-box from UBR2 in complex with HIFS peptide | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase UBR2, HIS-ILE-PHE-SER peptide, ... | | Authors: | Munoz-Escobar, J, Kozlov, G, Gehring, K. | | Deposit date: | 2016-09-19 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Bound Waters Mediate Binding of Diverse Substrates to a Ubiquitin Ligase.
Structure, 25, 2017
|
|
5TDB
 
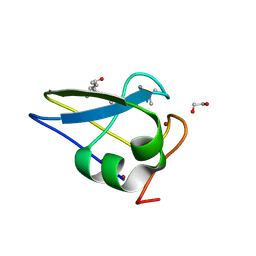 | | Crystal structure of the human UBR-box domain from UBR2 in complex with asymmetrically double methylated arginine peptide | | Descriptor: | 1,2-ETHANEDIOL, DA2-ILE-PHE-SER peptide, E3 ubiquitin-protein ligase UBR2, ... | | Authors: | Munoz-Escobar, J, Kozlov, G, Gehring, K. | | Deposit date: | 2016-09-19 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | Bound Waters Mediate Binding of Diverse Substrates to a Ubiquitin Ligase.
Structure, 25, 2017
|
|
5UM3
 
 | |
5V44
 
 | |
5VMD
 
 | | Crystal structure of UBR-box from UBR6 in a domain-swapping conformation | | Descriptor: | 1,2-ETHANEDIOL, F-box only protein 11, ZINC ION | | Authors: | Munoz-Escobar, J, Kozlov, G, Gehring, K. | | Deposit date: | 2017-04-27 | | Release date: | 2017-07-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Crystal structure of the UBR-box from UBR6/FBXO11 reveals domain swapping mediated by zinc binding.
Protein Sci., 26, 2017
|
|
5V45
 
 | |
5V46
 
 | | Crystal structure of the I113M, F270M, K291M, L308M mutant of SR1 domain of human sacsin | | Descriptor: | Sacsin | | Authors: | Menade, M, Kozlov, G, Gehring, K. | | Deposit date: | 2017-03-08 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of ubiquitin-like (Ubl) and Hsp90-like domains of sacsin provide insight into pathological mutations.
J. Biol. Chem., 293, 2018
|
|
5V47
 
 | | Crystal structure of the SR1 domain of lizard sacsin | | Descriptor: | Lizard sacsin, SULFATE ION | | Authors: | Pan, T, Menade, M, Kozlov, G, Gehring, K. | | Deposit date: | 2017-03-08 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structures of ubiquitin-like (Ubl) and Hsp90-like domains of sacsin provide insight into pathological mutations.
J. Biol. Chem., 293, 2018
|
|
5VXQ
 
 | | X-Ray crystallography structure of the parallel stranded duplex formed by 5-rA5-dA-rA5 | | Descriptor: | AMMONIUM ION, DNA/RNA (5'-R(*AP*AP*AP*AP*A)-D(P*A)-R(P*AP*AP*AP*AP*A)-3') | | Authors: | Xie, J, Chen, Y, Wei, X, Kozlov, G, Gehring, K. | | Deposit date: | 2017-05-23 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.002 Å) | | Cite: | Influence of nucleotide modifications at the C2' position on the Hoogsteen base-paired parallel-stranded duplex of poly(A) RNA.
Nucleic Acids Res., 45, 2017
|
|
