5C6E
 
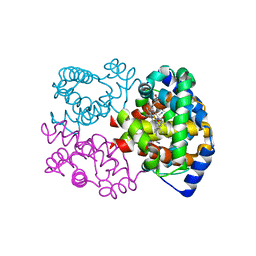 | | Joint X-ray/neutron structure of equine cyanomet hemoglobin in R state | | Descriptor: | CYANIDE ION, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Dajnowicz, S, Sean, S, Hanson, B.L, Fisher, S.Z, Langan, P, Kovalevsky, A.Y, Mueser, T.C. | | Deposit date: | 2015-06-22 | | Release date: | 2016-06-22 | | Last modified: | 2024-03-06 | | Method: | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | Cite: | Visualizing the Bohr effect in hemoglobin: neutron structure of equine cyanomethemoglobin in the R state and comparison with human deoxyhemoglobin in the T state.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5C8I
 
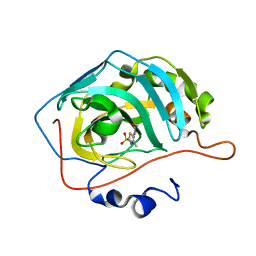 | | Joint X-ray/neutron structure of Human Carbonic Anhydrase II in complex with Methazolamide | | Descriptor: | Carbonic anhydrase 2, N-(3-methyl-5-sulfamoyl-1,3,4-thiadiazol-2(3H)-ylidene)acetamide, ZINC ION | | Authors: | Aggarwal, M, Kovalevsky, A.Y, Fisher, S.Z, McKenna, R. | | Deposit date: | 2015-06-25 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-06 | | Method: | NEUTRON DIFFRACTION (1.56 Å), X-RAY DIFFRACTION | | Cite: | Neutron structure of human carbonic anhydrase II in complex with methazolamide: mapping the solvent and hydrogen-bonding patterns of an effective clinical drug.
IUCrJ, 3, 2016
|
|
3NU3
 
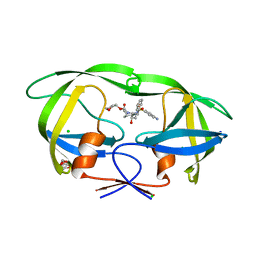 | | Wild Type HIV-1 Protease with Antiviral Drug Amprenavir | | Descriptor: | CHLORIDE ION, GLYCEROL, Protease, ... | | Authors: | Wang, Y.-F, Kovalevsky, A.Y, Weber, I.T. | | Deposit date: | 2010-07-06 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Amprenavir complexes with HIV-1 protease and its drug-resistant mutants altering hydrophobic clusters.
Febs J., 277, 2010
|
|
3NU4
 
 | | Crystal Structure of HIV-1 Protease Mutant V32I with Antiviral Drug Amprenavir | | Descriptor: | CHLORIDE ION, SODIUM ION, protease, ... | | Authors: | Wang, Y.-F, Kovalevsky, A.Y, Weber, I.T. | | Deposit date: | 2010-07-06 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Amprenavir complexes with HIV-1 protease and its drug-resistant mutants altering hydrophobic clusters.
Febs J., 277, 2010
|
|
7TUR
 
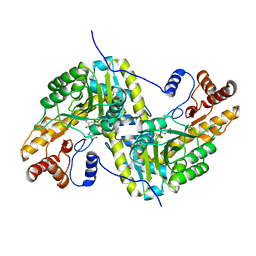 | | Joint X-ray/neutron structure of aspastate aminotransferase (AAT) in complex with pyridoxamine 5'-phosphate (PMP) | | Descriptor: | 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-AMINO]-2-METHYL-SUCCINIC ACID, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aspartate aminotransferase, ... | | Authors: | Drago, V.N, Kovalevsky, A.Y, Dajnowicz, S, Mueser, T.C. | | Deposit date: | 2022-02-03 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | Cite: | An N⋯H⋯N low-barrier hydrogen bond preorganizes the catalytic site of aspartate aminotransferase to facilitate the second half-reaction.
Chem Sci, 13, 2022
|
|
8EYS
 
 | |
5R42
 
 | | Crystal Structure of deuterated gamma-Chymotrypsin at pH 7.5, room temperature | | Descriptor: | IODIDE ION, gamma-Chymotrypsin, peptide SWPW, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
5R49
 
 | | Crystal Structure of gamma-Chymotrypsin at pH 5.6, cryo temperature | | Descriptor: | IODIDE ION, MALONATE ION, gamma-chymotrypsin, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
5R45
 
 | | Crystal Structure of gamma-Chymotrypsin at pH 7.5, cryo temperature | | Descriptor: | Chymotrypsinogen A, IODIDE ION, MALONATE ION, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
5R4C
 
 | | Crystal Structure of gamma-Chymotrypsin at pH 9, room temperature | | Descriptor: | IODIDE ION, SULFATE ION, gamma-chymotrypsin, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
5R48
 
 | | Crystal Structure of gamma-Chymotrypsin at pH 5.6, room temperature | | Descriptor: | IODIDE ION, SULFATE ION, gamma-chymotrypsin, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
5R4A
 
 | | Crystal Structure of deuterated gamma-Chymotrypsin at pH 9, room temperature | | Descriptor: | IODIDE ION, SULFATE ION, gamma-chymotrypsin, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
5R44
 
 | | Crystal Structure of gamma-Chymotrypsin at pH 7.5, room temperature | | Descriptor: | Chymotrypsinogen A, IODIDE ION, peptide SWPW, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
5R46
 
 | | Crystal Structure of deuterated gamma-Chymotrypsin at pH 5.6, room temperature | | Descriptor: | IODIDE ION, SULFATE ION, gamma-chymotrypsin, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
5R43
 
 | | Crystal Structure of deuterated gamma-Chymotrypsin at pH 7.5, cryo temperature | | Descriptor: | Chymotrypsinogen A, IODIDE ION, MALONIC ACID, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
5R4B
 
 | | Crystal Structure of deuterated gamma-Chymotrypsin at pH 9, cryo temperature | | Descriptor: | IODIDE ION, SULFATE ION, gamma-chymotrypsin, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
5R4D
 
 | | Crystal Structure of gamma-Chymotrypsin at pH 9, cryo temperature | | Descriptor: | IODIDE ION, SULFATE ION, gamma-chymotrypsin, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
5R47
 
 | | Crystal Structure of deuterated gamma-Chymotrypsin at pH 5.6, cryo temperature | | Descriptor: | IODIDE ION, MALONIC ACID, gamma-chymotrypsin, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
3I6O
 
 | | Crystal structure of wild type HIV-1 protease with macrocyclic inhibitor GRL-0216A | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(1S,2R)-1-benzyl-2-hydroxy-3-[(7E)-13-methoxy-1,1-dioxido-3,4,5,6,9,10-hexahydro-2H-11,1,2-benzoxathiazacyclotridecin-2-yl]propyl}carbamate, GLYCEROL, IODIDE ION, ... | | Authors: | Chumanevich, A.A, Wang, Y.-F, Kovalevsky, A.Y, Weber, I.T. | | Deposit date: | 2009-07-07 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Design, Synthesis, Protein-Ligand X-ray Structure, and Biological Evaluation of a Series of Novel Macrocyclic Human Immunodeficiency Virus-1 Protease Inhibitors to Combat Drug Resistance.
J.Med.Chem., 52, 2009
|
|
4QX5
 
 | | Neutron diffraction reveals hydrogen bonds critical for cGMP-selective activation: Insights for PKG agonist design | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, IODIDE ION, cGMP-dependent protein kinase 1 | | Authors: | Huang, G.Y, Gerlits, O.O, Blakeley, M.P, Sankaran, B, Kovalevsky, A.Y, Kim, C. | | Deposit date: | 2014-07-18 | | Release date: | 2014-11-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.318 Å) | | Cite: | Neutron Diffraction Reveals Hydrogen Bonds Critical for cGMP-Selective Activation: Insights for cGMP-Dependent Protein Kinase Agonist Design.
Biochemistry, 53, 2014
|
|
6XKF
 
 | | The crystal structure of 3CL MainPro of SARS-CoV-2 with oxidized Cys145 (Sulfenic acid cysteine). | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, CHLORIDE ION | | Authors: | Tan, K, Maltseva, N.I, Welk, L.F, Jedrzejczak, R.P, Coates, L, Kovalevskyi, A.Y, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-26 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of 3CL MainPro of SARS-CoV-2 with oxidized Cys145 (Sulfenic acid cysteine).
To Be Published
|
|
