1AD4
 
 | |
6FHS
 
 | | CryoEM Structure of INO80core | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Arp5, ... | | Authors: | Eustermann, S, Schall, K, Kostrewa, D, Strauss, M, Hopfner, K. | | Deposit date: | 2018-01-15 | | Release date: | 2018-04-25 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.754 Å) | | Cite: | Structural basis for ATP-dependent chromatin remodelling by the INO80 complex.
Nature, 556, 2018
|
|
6FML
 
 | | CryoEM Structure INO80core Nucleosome complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin related protein 5, ... | | Authors: | Eustermann, S, Schall, K, Kostrewa, D, Strauss, M, Hopfner, K. | | Deposit date: | 2018-01-31 | | Release date: | 2018-04-25 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.34 Å) | | Cite: | Structural basis for ATP-dependent chromatin remodelling by the INO80 complex.
Nature, 556, 2018
|
|
3N8B
 
 | | Crystal Structure of Borrelia burgdorferi Pur-alpha | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Uncharacterized protein | | Authors: | Graebsch, A, Roche, S, Kostrewa, D, Niessing, D. | | Deposit date: | 2010-05-28 | | Release date: | 2010-10-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Of bits and bugs--on the use of bioinformatics and a bacterial crystal structure to solve a eukaryotic repeat-protein structure.
Plos One, 5, 2010
|
|
3T2N
 
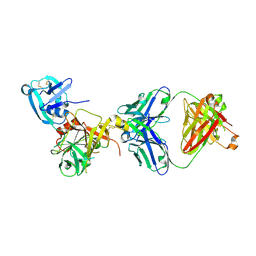 | | Human hepsin protease in complex with the Fab fragment of an inhibitory antibody | | Descriptor: | Antibody, Fab fragment, Heavy Chain, ... | | Authors: | Koschubs, T, Dengl, S, Duerr, H, Kaluza, K, Georges, G, Hartl, C, Jennewein, S, Lanzendoerfer, M, Auer, J, Stern, A, Huang, K.-S, Kostrewa, D, Ries, S, Hansen, S, Kohnert, U, Cramer, P, Mundigl, O. | | Deposit date: | 2011-07-22 | | Release date: | 2011-12-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Allosteric antibody inhibition of human hepsin protease.
Biochem.J., 442, 2012
|
|
3QT1
 
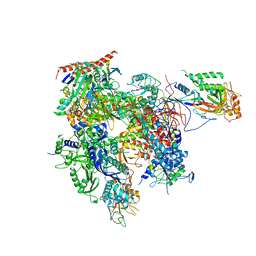 | | RNA polymerase II variant containing A Chimeric RPB9-C11 subunit | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Ruan, W, Lehmann, E, Thomm, M, Kostrewa, D, Cramer, P. | | Deposit date: | 2011-02-22 | | Release date: | 2011-03-23 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Evolution of two modes of intrinsic RNA polymerase transcript cleavage.
J.Biol.Chem., 286, 2011
|
|
3NM7
 
 | | Crystal Structure of Borrelia burgdorferi Pur-alpha | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Uncharacterized protein | | Authors: | Graebsch, A, Roche, S, Kostrewa, D, Niessing, D. | | Deposit date: | 2010-06-22 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Of Bits and Bugs - on the use of bioinformatics and a bacterial crystal structure to solve a eukaryotic repeat-protein structure.
Plos One, 5, 2010
|
|
6SKY
 
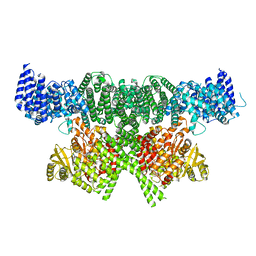 | | FAT and kinase domain of CtTel1 | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase Tel1 | | Authors: | Jansma, M, Eustermann, S.E, Kostrewa, D, Lammens, K, Hopfner, K.P. | | Deposit date: | 2019-08-16 | | Release date: | 2019-10-30 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Near-Complete Structure and Model of Tel1ATM from Chaetomium thermophilum Reveals a Robust Autoinhibited ATP State.
Structure, 28, 2020
|
|
6SL0
 
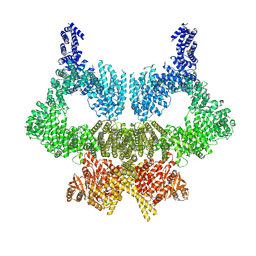 | | Complete CtTel1 dimer with C2 symmetry | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase Tel1 | | Authors: | Jansma, M, Eustermann, S.E, Kostrewa, D, Lammens, K, Hopfner, K.P. | | Deposit date: | 2019-08-16 | | Release date: | 2019-10-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Near-Complete Structure and Model of Tel1ATM from Chaetomium thermophilum Reveals a Robust Autoinhibited ATP State.
Structure, 28, 2020
|
|
6SL1
 
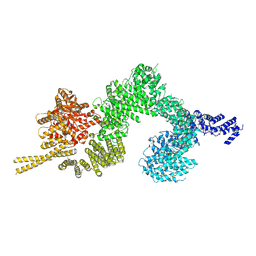 | | Structure of the open conformation of CtTel1 | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase Tel1 | | Authors: | Jansma, M, Eustermann, S.E, Kostrewa, D, Lammens, K, Hopfner, K.P. | | Deposit date: | 2019-08-16 | | Release date: | 2019-10-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Near-Complete Structure and Model of Tel1ATM from Chaetomium thermophilum Reveals a Robust Autoinhibited ATP State.
Structure, 28, 2020
|
|
6SKZ
 
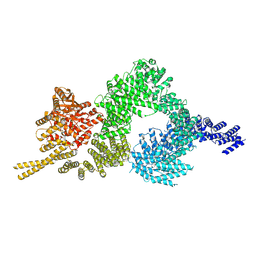 | | Structure of the closed conformation of CtTel1 | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase Tel1 | | Authors: | Jansma, M, Eustermann, S.E, Kostrewa, D, Lammens, K, Hopfner, K.P. | | Deposit date: | 2019-08-16 | | Release date: | 2019-10-30 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Near-Complete Structure and Model of Tel1ATM from Chaetomium thermophilum Reveals a Robust Autoinhibited ATP State.
Structure, 28, 2020
|
|
2AKF
 
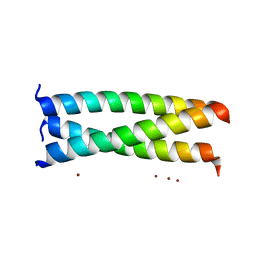 | | Crystal structure of the coiled-coil domain of coronin 1 | | Descriptor: | Coronin-1A, ZINC ION | | Authors: | Kammerer, R.A, Kostrewa, D, Progias, P, Honnappa, S, Avila, D, Lustig, A, Winkler, F.K, Pieters, J, Steinmetz, M.O. | | Deposit date: | 2005-08-03 | | Release date: | 2005-09-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | A conserved trimerization motif controls the topology of short coiled coils
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
8A5Q
 
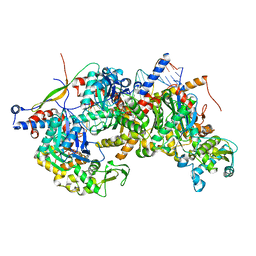 | | Structure of Arp4-Ies4-N-actin-Arp8-Ino80HSA subcomplex (A-module) of Chaetomium thermophilum INO80 on straight DNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin related protein 4 (Arp4), ... | | Authors: | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | Deposit date: | 2022-06-15 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
8ATF
 
 | | Nucleosome-bound Ino80 ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (226-MER), DNA (227-MER), ... | | Authors: | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | Deposit date: | 2022-08-23 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
8A5P
 
 | | Structure of Arp4-Ies4-N-actin-Arp8-Ino80HSA subcomplex (A-module) of Chaetomium thermophilum INO80 on curved DNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin related protein 4 (Arp4), ... | | Authors: | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | Deposit date: | 2022-06-15 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
8A5A
 
 | | Structure of Arp4-Ies4-N-actin-Arp8-Ino80HSA subcomplex (A-module) of INO80 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin-like protein ARP8, ... | | Authors: | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | Deposit date: | 2022-06-14 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
8AV6
 
 | | CryoEM structure of INO80 core nucleosome complex in closed grappler conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DASH complex subunit DAD4, ... | | Authors: | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | Deposit date: | 2022-08-26 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.68 Å) | | Cite: | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
8A5O
 
 | | Structure of Arp4-Ies4-N-actin-Arp8-Ino80HSA subcomplex (A-module) of S. cerevisiae INO80 | | Descriptor: | Actin, Actin-like protein ARP8, Actin-related protein 4, ... | | Authors: | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | Deposit date: | 2022-06-15 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
8A5D
 
 | | Structure of Arp4-Ies4-N-actin-Arp8-Ino80HSA subcomplex (A-module) of Chaetomium thermophilum INO80 | | Descriptor: | Actin, Actin related protein 4 (Arp4), Actin-related protein 8, ... | | Authors: | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | Deposit date: | 2022-06-14 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
3NFH
 
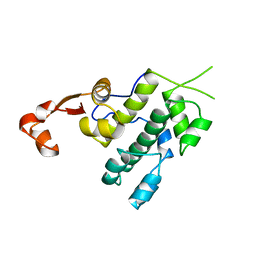 | | Crystal structure of tandem winged helix domain of RNA polymerase I subunit A49 (P4) | | Descriptor: | DNA-directed RNA polymerase I subunit RPA49 | | Authors: | Geiger, S.R, Lorenzen, K, Schreieck, A, Hanecker, P, Kostrewa, D, Heck, A.J.R, Cramer, P. | | Deposit date: | 2010-06-10 | | Release date: | 2010-09-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | RNA Polymerase I Contains a TFIIF-Related DNA-Binding Subcomplex.
Mol.Cell, 39, 2010
|
|
3OTX
 
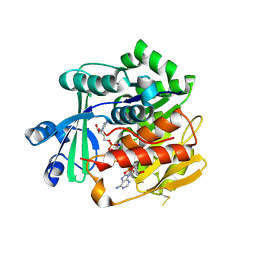 | | Crystal Structure of Trypanosoma brucei rhodesiense Adenosine Kinase Complexed with Inhibitor AP5A | | Descriptor: | Adenosine kinase, putative, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, ... | | Authors: | Kuettel, S, Greenwald, J, Kostrewa, D, Ahmed, S, Scapozza, L, Perozzo, R. | | Deposit date: | 2010-09-14 | | Release date: | 2011-06-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structures of T. b. rhodesiense Adenosine Kinase Complexed with Inhibitor and Activator: Implications for Catalysis and Hyperactivation
Plos Negl Trop Dis, 5, 2011
|
|
3NFF
 
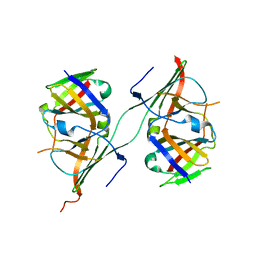 | | Crystal structure of extended Dimerization module of RNA polymerase I subcomplex A49/A34.5 | | Descriptor: | RNA polymerase I subunit A34.5, RNA polymerase I subunit A49 | | Authors: | Geiger, S.R, Lorenzen, K, Schreieck, A, Hanecker, P, Kostrewa, D, Heck, A.J.R, Cramer, P. | | Deposit date: | 2010-06-10 | | Release date: | 2010-09-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | RNA Polymerase I Contains a TFIIF-Related DNA-Binding Subcomplex.
Mol.Cell, 39, 2010
|
|
3NFI
 
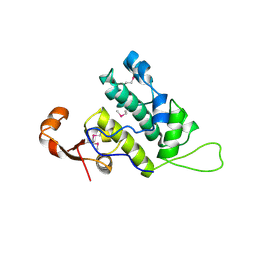 | | Crystal structure of tandem winged helix domain of RNA polymerase I subunit A49 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, DNA-directed RNA polymerase I subunit RPA49 | | Authors: | Geiger, S.R, Lorenzen, K, Schreieck, A, Hanecker, P, Kostrewa, D, Heck, A.J.R, Cramer, P. | | Deposit date: | 2010-06-10 | | Release date: | 2010-09-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | RNA Polymerase I Contains a TFIIF-Related DNA-Binding Subcomplex.
Mol.Cell, 39, 2010
|
|
1YNQ
 
 | | aldo-keto reductase AKR11C1 from Bacillus halodurans (holo form) | | Descriptor: | GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SODIUM ION, ... | | Authors: | Marquardt, T, Kostrewa, D, Winkler, F.K, Li, X.D. | | Deposit date: | 2005-01-25 | | Release date: | 2005-12-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High-resolution Crystal Structure of AKR11C1 from Bacillus halodurans: An NADPH-dependent 4-Hydroxy-2,3-trans-nonenal Reductase
J.Mol.Biol., 354, 2005
|
|
1YNP
 
 | | aldo-keto reductase AKR11C1 from Bacillus halodurans (apo form) | | Descriptor: | GLYCEROL, SODIUM ION, SULFATE ION, ... | | Authors: | Marquardt, T, Kostrewa, D, Winkler, F.K, Li, X.D. | | Deposit date: | 2005-01-25 | | Release date: | 2005-12-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | High-resolution Crystal Structure of AKR11C1 from Bacillus halodurans: An NADPH-dependent 4-Hydroxy-2,3-trans-nonenal Reductase
J.Mol.Biol., 354, 2005
|
|
