1S6Y
 
 | | 2.3A crystal structure of phospho-beta-glucosidase | | Descriptor: | 6-phospho-beta-glucosidase | | Authors: | Tereshko, V, Dementieva, I, Kim, Y, Collat, F, Joachimiak, A, Kossiakoff, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-01-28 | | Release date: | 2004-05-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | 2.3A CRYSTAL STRUCTURE OF PHOSPHO-BETA-GLUCOSIDASE, licH Gene Product from BACILLUS STEAROTHERMOPHILUS
To be Published
|
|
1S9U
 
 | | Atomic structure of a putative anaerobic dehydrogenase component | | Descriptor: | DI(HYDROXYETHYL)ETHER, SULFATE ION, putative component of anaerobic dehydrogenases | | Authors: | Qiu, Y, Zhang, R, Tereshko, V, Kim, Y, Collart, F, Joachimiak, A, Kossiakoff, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-05 | | Release date: | 2004-06-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | The 1.38 A crystal structure of DmsD protein from Salmonella typhimurium, a proofreading chaperone on the Tat pathway.
Proteins, 71, 2008
|
|
5VZ1
 
 | | Crystal structure of the Apo Antibody fragment (Fab) raised against C-terminal domain of Ebola nucleoprotein (EBOV, TAFV, BDBV strains) | | Descriptor: | Apo Antibody Fab Heavy Chain, Apo Antibody Fab Light Chain, ZINC ION | | Authors: | Radwanska, M.J, Derewenda, U, Kossiakoff, A, Derewenda, Z.S. | | Deposit date: | 2017-05-26 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.112 Å) | | Cite: | Crystal structure of the Apo Antibody fragment (Fab) raised against C-terminal domain of Ebola nucleoprotein (EBOV, TAFV, BDBV strains)
To Be Published
|
|
5VKD
 
 | | Crystal structure of C-terminal domain of Ebola (Bundibugyo) nucleoprotein in complex with Fab fragment | | Descriptor: | Fab Heavy Chain, Fab light chain, Nucleoprotein | | Authors: | Radwanska, M.J, Derewenda, U, Kossiakoff, A, Derewenda, Z.S. | | Deposit date: | 2017-04-21 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | The structure of the C-terminal domain of the nucleoprotein from the Bundibugyo strain of the Ebola virus in complex with a pan-specific synthetic Fab.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
3PJS
 
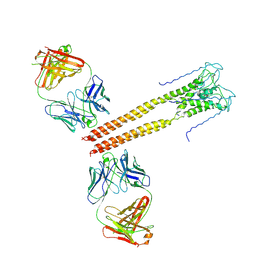 | | Mechanism of Activation Gating in the Full-Length KcsA K+ Channel | | Descriptor: | FAB heavy chain, FAB light chain, Voltage-gated potassium channel | | Authors: | Uysal, S, Cuello, L.G, Kossiakoff, A, Perozo, E. | | Deposit date: | 2010-11-10 | | Release date: | 2011-07-06 | | Last modified: | 2011-08-03 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Mechanism of activation gating in the full-length KcsA K+ channel.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1R4V
 
 | | 1.9A crystal structure of protein AQ328 from Aquifex aeolicus | | Descriptor: | CACODYLATE ION, Hypothetical protein AQ_328, ZINC ION | | Authors: | Qiu, Y, Tereshko, V, Kim, Y, Zhang, R, Collart, F, Joachimiak, A, Kossiakoff, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-10-08 | | Release date: | 2004-03-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of Aq_328 from the hyperthermophilic bacteria Aquifex aeolicus shows an ancestral histone fold.
Proteins, 62, 2006
|
|
7KEO
 
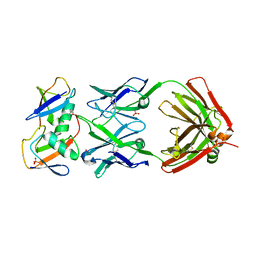 | | Crystal structure of K29-linked di-ubiquitin in complex with synthetic antigen binding fragment | | Descriptor: | PHOSPHATE ION, Synthetic antigen binding fragment, heavy chain, ... | | Authors: | Yu, Y, Zheng, Q, Erramilli, S, Pan, M, Kossiakoff, A, Liu, L, Zhao, M. | | Deposit date: | 2020-10-11 | | Release date: | 2021-07-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | K29-linked ubiquitin signaling regulates proteotoxic stress response and cell cycle.
Nat.Chem.Biol., 17, 2021
|
|
3EFF
 
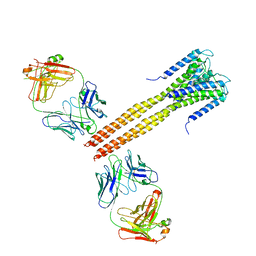 | | The Crystal Structure of Full-Length KcsA in its Closed Conformation | | Descriptor: | FAB, Voltage-gated potassium channel | | Authors: | Uysal, S, Vasquez, V, Tereshko, T, Esaki, K, Fellouse, F.A, Sidhu, S.S, Koide, S, Perozo, E, Kossiakoff, A. | | Deposit date: | 2008-09-08 | | Release date: | 2009-04-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structure of full-length KcsA in its closed conformation.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1U14
 
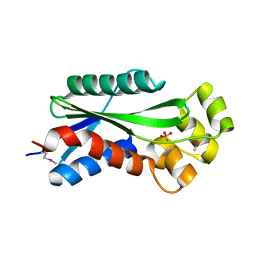 | | The crystal structure of hypothetical UPF0244 protein yjjX at resolution 1.68 Angstrom | | Descriptor: | Hypothetical UPF0244 protein yjjX, PHOSPHATE ION | | Authors: | Qiu, Y, Kim, Y, Cuff, M, Collart, F, Joachimiak, A, Kossiakoff, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-07-14 | | Release date: | 2004-09-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The crystal structure of hypothetical UPF0244 protein yjjX at resolution 1.68 Angstrom
To be Published
|
|
3EFD
 
 | | The crystal structure of the cytoplasmic domain of KcsA | | Descriptor: | FabH, FabL, KcsA | | Authors: | Uysal, S, Vasquez, V, Tereshko, V, Esaki, K, Fellouse, F.A, Sidhu, S.S, Koide, S, Perozo, E, Kossiakoff, A. | | Deposit date: | 2008-09-08 | | Release date: | 2009-04-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of full-length KcsA in its closed conformation.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1Y7R
 
 | | 1.7 A Crystal Structure of Protein of Unknown Function SA2161 from Meticillin-Resistant Staphylococcus aureus, Probable Acetyltransferase | | Descriptor: | PHOSPHATE ION, hypothetical protein SA2161 | | Authors: | Qiu, Y, Zhang, R, Collart, F, Holzle, D, Joachimiak, A, Kossiakoff, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-12-09 | | Release date: | 2005-01-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.7A Crystal Structure of Hypothetical Protein SA2161 from Meticillin-Resistant Staphylococcus aureus
To be Published
|
|
2ARH
 
 | | Crystal Structure of a Protein of Unknown Function AQ1966 from Aquifex aeolicus VF5 | | Descriptor: | CALCIUM ION, SELENIUM ATOM, SULFATE ION, ... | | Authors: | Qiu, Y, Kim, Y, Yang, X, Collart, F, Joachimiak, A, Kossiakoff, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-08-19 | | Release date: | 2005-10-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Crystal Structure of a Hypothetical Protein Aq_1966 from Aquifex aeolicus VF5
To be Published
|
|
1X87
 
 | | 2.4A X-ray structure of Urocanase protein complexed with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Urocanase protein | | Authors: | Tereshko, V, Zaborske, J, Gilbreth, R, Dementieva, I, Collart, F, Joachimiak, A, Kossiakoff, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-17 | | Release date: | 2004-10-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4A X-ray structure of Urocanase protein complexed with NAD
To be Published
|
|
2H1J
 
 | | 3.1 A X-ray structure of putative Oligoendopeptidase F: Crystals grown by microfluidic seeding | | Descriptor: | Oligoendopeptidase F, ZINC ION | | Authors: | Gerdts, C.J, Tereshko, V, Dementieva, I, Collart, F, Joachimiak, A, Kossiakoff, A, Ismagilov, R.F, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2006-05-16 | | Release date: | 2006-06-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Time-Controlled Microfluidic Seeding in nL-Volume Droplets To Separate Nucleation and Growth Stages of Protein Crystallization.
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
2H1N
 
 | | 3.0 A X-ray structure of putative oligoendopeptidase F: crystals grown by vapor diffusion technique | | Descriptor: | Oligoendopeptidase F, UNKNOWN LIGAND, ZINC ION | | Authors: | Gerdts, C.J, Tereshko, V, Dementieva, I, Collart, F, Joachimiak, A, Kossiakoff, A, Ismagilov, R.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-05-16 | | Release date: | 2006-06-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Time-Controlled Microfluidic Seeding in nL-Volume Droplets To Separate Nucleation and Growth Stages of Protein Crystallization.
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
7MDJ
 
 | | The structure of KcsA in complex with a synthetic Fab | | Descriptor: | Fab heavy chain, Fab light chain, POTASSIUM ION, ... | | Authors: | Rohaim, A, Slezak, T, Blackowicz, L, Kossiakoff, A, Roux, B. | | Deposit date: | 2021-04-05 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Engineering of a synthetic antibody fragment for structural and functional studies of K+ channels.
J.Gen.Physiol., 154, 2022
|
|
8TNJ
 
 | | Cryo-EM structure of HLA-B*73:01 bound to a 9mer peptide and two Fabs | | Descriptor: | 9mer peptide,Beta-2-microglobulin,MHC class I antigen chimera, B.1 Fab heavy chain, B.1 Fab light chain, ... | | Authors: | Ross, P, Adams, E.J, Lodwick, J, Zhao, M, Slezak, T, Kossiakoff, A. | | Deposit date: | 2023-08-02 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of HLA-B*73:01 bound to a 9mer peptide and two Fabs
To Be Published
|
|
