1WVG
 
 | | Structure of CDP-D-glucose 4,6-dehydratase from Salmonella typhi | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CDP-glucose 4,6-dehydratase, CYTIDINE-5'-DIPHOSPHO-BETA-D-XYLOSE | | Authors: | Koropatkin, N.M, Holden, H.M. | | Deposit date: | 2004-12-15 | | Release date: | 2005-01-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of CDP-D-glucose 4,6-dehydratase from Salmonella typhi complexed with CDP-D-xylose.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4UAC
 
 | | EUR_01830 with acarbose | | Descriptor: | 1,2-ETHANEDIOL, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Carbohydrate ABC transporter substrate-binding protein, ... | | Authors: | Koropatkin, N.M, Orlovsky, N.I. | | Deposit date: | 2014-08-08 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular details of a starch utilization pathway in the human gut symbiont Eubacterium rectale.
Mol.Microbiol., 95, 2015
|
|
4UA8
 
 | |
2G29
 
 | |
1ORR
 
 | |
2PT2
 
 | | Structure of FutA1 with Iron(II) | | Descriptor: | 1,2-ETHANEDIOL, FE (II) ION, Iron transport protein, ... | | Authors: | Koropatkin, N.M, Randich, A.M, Bhattachryya-Pakrasi, M, Pakrasi, H.B, Smith, T.J. | | Deposit date: | 2007-05-07 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the iron-binding protein, FutA1, from Synechocystis 6803.
J.Biol.Chem., 282, 2007
|
|
2PT1
 
 | | FutA1 Synechocystis PCC 6803 | | Descriptor: | Iron transport protein, SULFATE ION | | Authors: | Koropatkin, N.M, Randich, A.M, Bhattachryya-Pakrasi, M, Pakrasi, H.B, Smith, T.J. | | Deposit date: | 2007-05-07 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the iron-binding protein, FutA1, from Synechocystis 6803.
J.Biol.Chem., 282, 2007
|
|
1TZF
 
 | |
3CKC
 
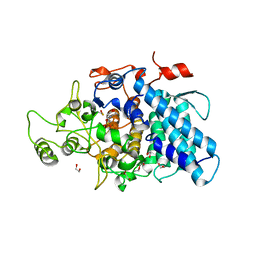 | | B. thetaiotaomicron SusD | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Koropatkin, N.M, Martens, E.C, Gordon, J.I, Smith, T.J. | | Deposit date: | 2008-03-14 | | Release date: | 2008-04-01 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices.
Structure, 16, 2008
|
|
1WVC
 
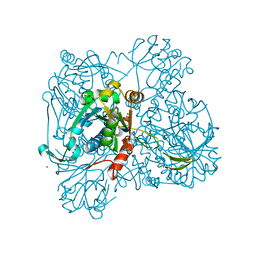 | | alpha-D-glucose-1-phosphate cytidylyltransferase complexed with CTP | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, Glucose-1-phosphate cytidylyltransferase, MAGNESIUM ION, ... | | Authors: | Koropatkin, N.M, Cleland, W.W, Holden, H.M. | | Deposit date: | 2004-12-14 | | Release date: | 2005-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Kinetic and structural analysis of alpha-D-Glucose-1-phosphate cytidylyltransferase from Salmonella typhi.
J.Biol.Chem., 280, 2005
|
|
3CKB
 
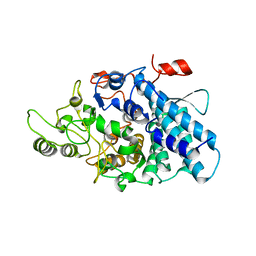 | | B. thetaiotaomicron SusD with maltotriose | | Descriptor: | CALCIUM ION, SusD, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Koropatkin, N.M, Martens, E.C, Gordon, J.I, Smith, T.J. | | Deposit date: | 2008-03-14 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices.
Structure, 16, 2008
|
|
3EHM
 
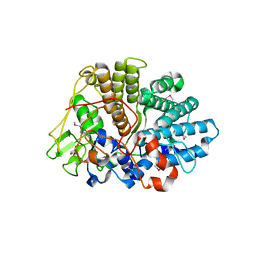 | | Structure of BT1043 | | Descriptor: | 1,2-ETHANEDIOL, SusD homolog | | Authors: | Koropatkin, N.M, Smith, T.J. | | Deposit date: | 2008-09-13 | | Release date: | 2009-05-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a SusD homologue, BT1043, involved in mucin O-glycan utilization in a prominent human gut symbiont.
Biochemistry, 48, 2009
|
|
5VOL
 
 | | Bacint_04212 ferulic acid esterase | | Descriptor: | CALCIUM ION, ISOPROPYL ALCOHOL, Putative esterase | | Authors: | Koropatkin, N.M, Cann, I, Mackie, R.I. | | Deposit date: | 2017-05-03 | | Release date: | 2017-07-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Biochemical and Structural Analyses of Two Cryptic Esterases in Bacteroides intestinalis and their Synergistic Activities with Cognate Xylanases.
J. Mol. Biol., 429, 2017
|
|
5WH8
 
 | | Cellulase Cel5C_n | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, PENTAETHYLENE GLYCOL, ... | | Authors: | Koropatkin, N.M, Pope, P.B, Naas, A.E. | | Deposit date: | 2017-07-15 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | "Candidatus Paraporphyromonas polyenzymogenes" encodes multi-modular cellulases linked to the type IX secretion system.
Microbiome, 6, 2018
|
|
5J5U
 
 | | Fjoh_4561 chitin-binding protein | | Descriptor: | RagB/SusD domain protein | | Authors: | Koropatkin, N.M. | | Deposit date: | 2016-04-03 | | Release date: | 2017-02-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A polysaccharide utilization locus from Flavobacterium johnsoniae enables conversion of recalcitrant chitin.
Biotechnol Biofuels, 9, 2016
|
|
5J90
 
 | |
3CK7
 
 | | B. thetaiotaomicron SusD with alpha-cyclodextrin | | Descriptor: | CALCIUM ION, Cyclohexakis-(1-4)-(alpha-D-glucopyranose), SusD | | Authors: | Koropatkin, N.M, Martens, E.C, Gordon, J.I, Smith, T.J. | | Deposit date: | 2008-03-14 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices.
Structure, 16, 2008
|
|
3CK8
 
 | | B. thetaiotaomicron SusD with beta-cyclodextrin | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), ... | | Authors: | Koropatkin, N.M, Martens, E.C, Gordon, J.I, Smith, T.J. | | Deposit date: | 2008-03-14 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices.
Structure, 16, 2008
|
|
3CK9
 
 | | B. thetaiotaomicron SusD with maltoheptaose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, SusD, ... | | Authors: | Koropatkin, N.M, Martens, E.C, Gordon, J.I, Smith, T.J. | | Deposit date: | 2008-03-14 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices.
Structure, 16, 2008
|
|
4FEM
 
 | | Structure of SusE with alpha-cyclodextrin | | Descriptor: | 1,2-ETHANEDIOL, Cyclohexakis-(1-4)-(alpha-D-glucopyranose), Outer membrane protein SusE | | Authors: | Koropatkin, N.M, Cameron, E.A, Martens, E.C. | | Deposit date: | 2012-05-30 | | Release date: | 2012-08-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Multidomain Carbohydrate-binding Proteins Involved in Bacteroides thetaiotaomicron Starch Metabolism.
J.Biol.Chem., 287, 2012
|
|
4FCH
 
 | | Crystal Structure SusE from Bacteroides thetaiotaomicron with maltoheptaose | | Descriptor: | 1,2-ETHANEDIOL, Outer membrane protein SusE, SULFATE ION, ... | | Authors: | Koropatkin, N.M, Cameron, E.A, Martens, E.C. | | Deposit date: | 2012-05-24 | | Release date: | 2012-08-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Multidomain Carbohydrate-binding Proteins Involved in Bacteroides thetaiotaomicron Starch Metabolism.
J.Biol.Chem., 287, 2012
|
|
4FE9
 
 | | Crystal Structure of SusF from Bacteroides thetaiotaomicron | | Descriptor: | 1,2-ETHANEDIOL, Cyclic alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose, GLYCEROL, ... | | Authors: | Koropatkin, N.M, Cameron, E.A, Martens, E.C. | | Deposit date: | 2012-05-29 | | Release date: | 2012-08-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Multidomain Carbohydrate-binding Proteins Involved in Bacteroides thetaiotaomicron Starch Metabolism.
J.Biol.Chem., 287, 2012
|
|
3F11
 
 | | Structure of futa1 with iron(III) | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, Iron transport protein, ... | | Authors: | Koropatkin, N.M, Randich, A.M, Bhattachryya-Pakrasi, M, Pakrasi, H.B, Smith, T.J. | | Deposit date: | 2008-10-27 | | Release date: | 2008-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the iron-binding protein, FutA1, from Synechocystis 6803
J.Biol.Chem., 282, 2007
|
|
3K8L
 
 | |
3K8M
 
 | | Crystal structure of SusG with acarbose | | Descriptor: | 1,2-ETHANEDIOL, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Koropatkin, N.M, Smith, T.J. | | Deposit date: | 2009-10-14 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | SusG: A Unique Cell-Membrane-Associated alpha-Amylase from a Prominent Human Gut Symbiont Targets Complex Starch Molecules.
Structure, 18, 2010
|
|
