3UX2
 
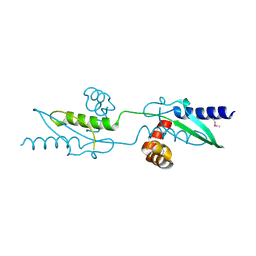 | |
7LCZ
 
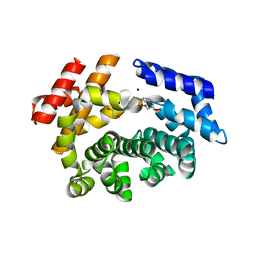 | | Crystal structure of the ARM domain from Drosophila SARM1 in complex with NMN | | Descriptor: | 1,2-ETHANEDIOL, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, Isoform B of NAD(+) hydrolase sarm1, ... | | Authors: | Gu, W, Nanson, J.D, Luo, Z, Jia, X, Manik, M.K, Ve, T, Kobe, B. | | Deposit date: | 2021-01-12 | | Release date: | 2021-03-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | SARM1 is a metabolic sensor activated by an increased NMN/NAD + ratio to trigger axon degeneration.
Neuron, 109, 2021
|
|
3ZIQ
 
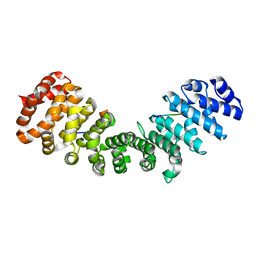 | | minor-site specific NLS (B6) | | Descriptor: | B6NLS, IMPORTIN SUBUNIT ALPHA-2 | | Authors: | Chang, C.-W, Counago, R.M, Williams, S.J, Kobe, B. | | Deposit date: | 2013-01-10 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Distinctive Conformation of Minor Site-Specific Nuclear Localization Signals Bound to Importin-Alpha
Traffic, 14, 2013
|
|
3ZIP
 
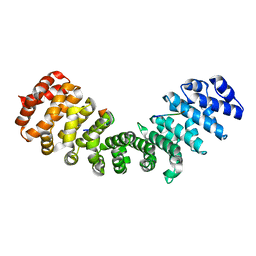 | | minor-site specific NLS (A58) | | Descriptor: | A58NLS, IMPORTIN SUBUNIT ALPHA-2 | | Authors: | Chang, C.-W, Counago, R.M, Williams, S.J, Kobe, B. | | Deposit date: | 2013-01-10 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Distinctive Conformation of Minor Site-Specific Nuclear Localization Signals Bound to Importin-Alpha
Traffic, 14, 2013
|
|
7JJ8
 
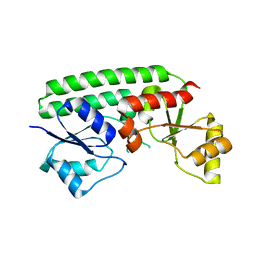 | | Crystal structure of the Zn(II)-bound ZnuA-like domain of Streptococcus pneumoniae AdcA | | Descriptor: | ZINC ION, Zinc-binding lipoprotein AdcA | | Authors: | Luo, Z, More, J.R, Kobe, B, McDevitt, C.A. | | Deposit date: | 2020-07-24 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A Trap-Door Mechanism for Zinc Acquisition by Streptococcus pneumoniae AdcA.
Mbio, 12, 2021
|
|
7LVP
 
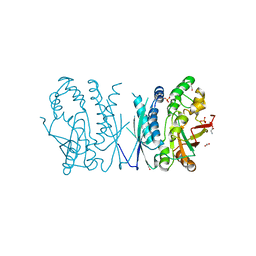 | | Cryptococcus neoformans AIR synthetase | | Descriptor: | 1,2-ETHANEDIOL, AIR synthase, GLYCINE, ... | | Authors: | Chua, S.M.H, Luo, Z, Lim, B.Y.J, Kobe, B, Fraser, J.A. | | Deposit date: | 2021-02-26 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural features of Cryptococcus neoformans bifunctional GAR/AIR synthetase may present novel antifungal drug targets.
J.Biol.Chem., 297, 2021
|
|
7LVO
 
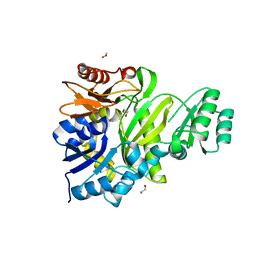 | | Cryptococcus neoformans GAR synthetase | | Descriptor: | 1,2-ETHANEDIOL, phosphoribosyl-glycinamide (GAR) synthetase | | Authors: | Chua, S.M.H, Luo, Z, Lim, B.Y.J, Kobe, B, Fraser, J.A. | | Deposit date: | 2021-02-26 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural features of Cryptococcus neoformans bifunctional GAR/AIR synthetase may present novel antifungal drug targets.
J.Biol.Chem., 297, 2021
|
|
4B8P
 
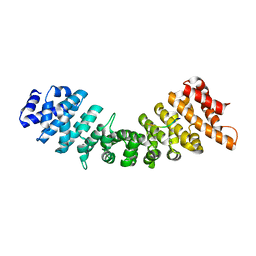 | | rImp_alpha_A89NLS | | Descriptor: | A89NLS, IMPORTIN SUBUNIT ALPHA-1A | | Authors: | Chang, C.-W, Counago, R.L.M, Williams, S.J, Boden, M, Kobe, B. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Rice Importin-Alpha and Structural Basis of its Interaction with Plant-Specific Nuclear Localization Signals.
Plant Cell, 24, 2012
|
|
4B8O
 
 | | rImp_alpha_SV40TAgNLS | | Descriptor: | IMPORTIN SUBUNIT ALPHA-1A, SV40TAGNLS | | Authors: | Chang, C.-W, Counago, R.L.M, Williams, S.J, Boden, M, Kobe, B. | | Deposit date: | 2012-08-28 | | Release date: | 2013-01-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.084 Å) | | Cite: | Crystal Structure of Rice Importin-Alpha and Structural Basis of its Interaction with Plant-Specific Nuclear Localization Signals.
Plant Cell, 24, 2012
|
|
3UX3
 
 | | Crystal Structure of Domain-Swapped Fam96a minor dimer | | Descriptor: | ACETATE ION, MIP18 family protein FAM96A, ZINC ION | | Authors: | Chen, K.-E, Kobe, B, Martin, J.L. | | Deposit date: | 2011-12-03 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The mammalian DUF59 protein Fam96a forms two distinct types of domain-swapped dimer.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
7LCY
 
 | | Crystal structure of the ligand-free ARM domain from Drosophila SARM1 | | Descriptor: | Isoform B of NAD(+) hydrolase sarm1 | | Authors: | Gu, W, Nanson, J.D, Luo, Z, McGuinness, H.Y, Manik, M.K, Jia, X, Ve, T, Kobe, B. | | Deposit date: | 2021-01-12 | | Release date: | 2021-03-10 | | Last modified: | 2021-04-21 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | SARM1 is a metabolic sensor activated by an increased NMN/NAD + ratio to trigger axon degeneration.
Neuron, 109, 2021
|
|
4DWN
 
 | | Crystal Structure of Human BinCARD CARD | | Descriptor: | Bcl10-interacting CARD protein, SULFATE ION | | Authors: | Chen, K.-E, Kobe, B, Martin, J.L. | | Deposit date: | 2012-02-26 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | The structure of the caspase recruitment domain of BinCARD reveals that all three cysteines can be oxidized.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3ZK8
 
 | | CRYSTAL STRUCTURE OF PNEUMOCOCCAL SURFACE ANTIGEN PSAA E205Q IN THE METAL-FREE, OPEN STATE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE ABC TRANSPORTER SUBSTRATE-BINDING LIPOPROTEIN | | Authors: | Counago, R.M, Ween, M.P, Bajaj, M, Zuegg, J, Cooper, M.A, McEwan, A.G, Paton, J.C, Kobe, B, McDevitt, C.A. | | Deposit date: | 2013-01-22 | | Release date: | 2013-11-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Imperfect coordination chemistry facilitates metal ion release in the Psa permease.
Nat. Chem. Biol., 10, 2014
|
|
4C0M
 
 | | Crystal Structure of the N terminal domain of wild type TRIF (TIR- domain-containing adapter-inducing interferon-beta) | | Descriptor: | TIR DOMAIN-CONTAINING ADAPTER MOLECULE 1 | | Authors: | Ullah, M.O, Ve, T, Mangan, M, Alaidarous, M, Sweet, M.J, Mansell, A, Kobe, B. | | Deposit date: | 2013-08-05 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Tlr Signalling Adaptor Trif/Ticam-1 Has an N-Terminal Helical Domain with Structural Similarity to Ifit Proteins
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4C6S
 
 | | Crystal structure of the TIR domain from the Arabidopsis Thaliana disease resistance protein RRS1 | | Descriptor: | GLYCEROL, PROBABLE WRKY TRANSCRIPTION FACTOR 52, SODIUM ION, ... | | Authors: | Wan, L, Williams, S.J, Sohn, K.H, Bernoux, M, Ma, Y, Segonzac, C, Ve, T, Sarris, P, Ericsson, D.J, Saucet, S.B, Zhang, X, Parker, J, Dodds, P.N, Jones, J.D.G, Kobe, B. | | Deposit date: | 2013-09-19 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Structural Basis for Assembly and Function of a Heterodimeric Plant Immune Receptor.
Science, 344, 2014
|
|
7L6W
 
 | | SFX structure of the MyD88 TIR domain higher-order assembly | | Descriptor: | Myeloid differentiation primary response protein MyD88 | | Authors: | Clabbers, M.T.B, Holmes, S, Muusse, T.W, Vajjhala, P, Thygesen, S.J, Malde, A.K, Hunter, D.J.B, Croll, T.I, Flueckiger, L, Nanson, J.D, Rahaman, M.H, Aquila, A, Hunter, M.S, Liang, M, Yoon, C.H, Zhao, J, Zatsepin, N.A, Abbey, B, Sierecki, E, Gambin, Y, Stacey, K.J, Darmanin, C, Kobe, B, Xu, H, Ve, T. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | MyD88 TIR domain higher-order assembly interactions revealed by microcrystal electron diffraction and serial femtosecond crystallography.
Nat Commun, 12, 2021
|
|
3UKX
 
 | | Mouse importin alpha: Bimax2 peptide complex | | Descriptor: | Bimax2 peptide, Importin subunit alpha-2 | | Authors: | Marfori, M, Forwood, J.K, Lonhienne, T.G, Kobe, B. | | Deposit date: | 2011-11-10 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of High-Affinity Nuclear Localization Signal Interactions with Importin-alpha
Traffic, 13, 2012
|
|
4BSX
 
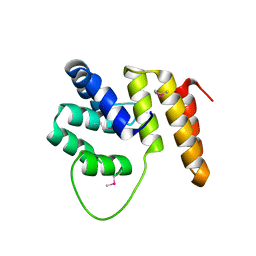 | | Crystal Structure of the N terminal domain of TRIF (TIR-domain- containing adapter-inducing interferon-beta) | | Descriptor: | TIR DOMAIN-CONTAINING ADAPTER MOLECULE 1 | | Authors: | Ullah, M.O, Ve, T, Mangan, M, Alaidarous, M, Sweet, M.J, Mansell, A, Kobe, B. | | Deposit date: | 2013-06-12 | | Release date: | 2013-12-11 | | Last modified: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (2.229 Å) | | Cite: | The Tlr Signalling Adaptor Trif/Ticam-1 Has an N-Terminal Helical Domain with Structural Similarity to Ifit Proteins
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4AF0
 
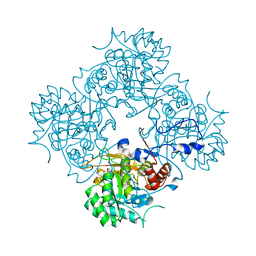 | | Crystal structure of cryptococcal inosine monophosphate dehydrogenase | | Descriptor: | INOSINE-5'-MONOPHOSPHATE DEHYDROGENASE, INOSINIC ACID, MYCOPHENOLIC ACID, ... | | Authors: | Valkov, E, Stamp, A, Morrow, C.A, Kobe, B, Fraser, J.A. | | Deposit date: | 2012-01-15 | | Release date: | 2012-10-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | De Novo GTP Biosynthesis is Critical for Virulence of the Fungal Pathogen Cryptococcus Neoformans
Plos Pathog., 8, 2012
|
|
3UL0
 
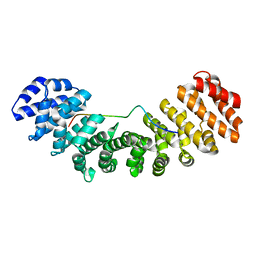 | | Mouse importin alpha: mouse CBP80Y8D cNLS complex | | Descriptor: | Importin subunit alpha-2, Nuclear cap-binding protein subunit 1 | | Authors: | Marfori, M, Forwood, J.K, Lonhienne, T.G, Kobe, B. | | Deposit date: | 2011-11-10 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of High-Affinity Nuclear Localization Signal Interactions with Importin-alpha
Traffic, 13, 2012
|
|
3UKW
 
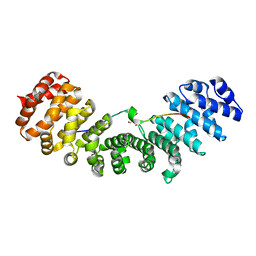 | | Mouse importin alpha: Bimax1 peptide complex | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bimax1 peptide, Importin subunit alpha-2 | | Authors: | Marfori, M, Forwood, J.K, Lonhienne, T.G, Kobe, B. | | Deposit date: | 2011-11-10 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of High-Affinity Nuclear Localization Signal Interactions with Importin-alpha
Traffic, 13, 2012
|
|
3UL1
 
 | | Mouse importin alpha: nucleoplasmin cNLS peptide complex | | Descriptor: | Importin subunit alpha-2, Nucleoplasmin | | Authors: | Marfori, M, Forwood, J.K, Lonhienne, T.G, Kobe, B. | | Deposit date: | 2011-11-10 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of High-Affinity Nuclear Localization Signal Interactions with Importin-alpha
Traffic, 13, 2012
|
|
3UKZ
 
 | | Mouse importin alpha: mouse CBP80 cNLS complex | | Descriptor: | Importin subunit alpha-2, Nuclear cap-binding protein subunit 1 | | Authors: | Marfori, M, Forwood, J.K, Lonhienne, T.G, Kobe, B. | | Deposit date: | 2011-11-10 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of High-Affinity Nuclear Localization Signal Interactions with Importin-alpha
Traffic, 13, 2012
|
|
3UKY
 
 | | Mouse importin alpha: yeast CBP80 cNLS complex | | Descriptor: | Importin subunit alpha-2, Nuclear cap-binding protein complex subunit 1 | | Authors: | Marfori, M, Forwood, J.K, Lonhienne, T.G, Kobe, B. | | Deposit date: | 2011-11-10 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of High-Affinity Nuclear Localization Signal Interactions with Importin-alpha
Traffic, 13, 2012
|
|
3ZLF
 
 | | Structure of group A Streptococcal enolase K312A mutant | | Descriptor: | ENOLASE, PHOSPHATE ION | | Authors: | Cork, A.J, Ericsson, D.J, Law, R.H.P, Casey, L.W, Valkov, E, Bertozzi, C, Stamp, A, Aquilina, J.A, Whisstock, J.C, Walker, M.J, Kobe, B. | | Deposit date: | 2013-01-31 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Stability of the Octameric Structure Affects Plasminogen-Binding Capacity of Streptococcal Enolase.
Plos One, 10, 2015
|
|
