2DFL
 
 | | Crystal structure of left-handed RadA filament | | Descriptor: | DNA repair and recombination protein radA | | Authors: | Chen, L.T, Ko, T.P, Wang, T.F, Wang, A.H.J. | | Deposit date: | 2006-03-02 | | Release date: | 2007-01-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the left-handed archaeal RadA helical filament: identification of a functional motif for controlling quaternary structures and enzymatic functions of RecA family proteins
Nucleic Acids Res., 35, 2007
|
|
2DH4
 
 | | Geranylgeranyl pyrophosphate synthase | | Descriptor: | MAGNESIUM ION, YPL069C | | Authors: | Chang, T.-H, Guo, R.-T, Ko, T.-P, Wang, A.H, Liang, P.-H. | | Deposit date: | 2006-03-22 | | Release date: | 2006-04-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structure of Type-III Geranylgeranyl Pyrophosphate Synthase from Saccharomyces cerevisiae and the Mechanism of Product Chain Length Determination.
J.Biol.Chem., 281, 2006
|
|
5ITJ
 
 | | The structure of histone-like protein | | Descriptor: | AbrB family transcriptional regulator, SULFATE ION, TETRAETHYLENE GLYCOL | | Authors: | Lin, B.L, Chen, C.Y, Huang, C.H, Ko, T.P, Chiang, C.H, Lin, K.F, Chang, Y.C, Lin, P.Y, Tsai, H.H.G, Wang, A.H.J. | | Deposit date: | 2016-03-17 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | The Arginine Pairs and C-Termini of the Sso7c4 from Sulfolobus solfataricus Participate in Binding and Bending DNA.
PLoS ONE, 12, 2017
|
|
2DTN
 
 | | Crystal structure of Helicobacter pylori undecaprenyl pyrophosphate synthase complexed with pyrophosphate | | Descriptor: | DIPHOSPHATE, undecaprenyl pyrophosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Chen, C.L, Ko, T.P, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2006-07-13 | | Release date: | 2007-06-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Biochemical characterization, crystal structure, and inhibitors of Helicobacter pylori undecaprenyl pyrophosphate synthase
To be Published
|
|
5XFT
 
 | |
2ZX9
 
 | | alpha-L-fucosidase complexed with inhibitor, B4 | | Descriptor: | (2S)-2-cyclopentyl-2-phenyl-N-{[(2R,3R,4R,5R,6S)-3,4,5-trihydroxy-6-methylpiperidin-2-yl]methyl}ethanamide, Alpha-L-fucosidase, putative | | Authors: | Wu, H.-J, Ko, T.-P, Ho, C.-W, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2008-12-22 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural basis of alpha-fucosidase inhibition by iminocyclitols with K(i) values in the micro- to picomolar range.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
3AMD
 
 | | Crystal structures of Thermotoga maritima Cel5A, apo form and tetramer/au | | Descriptor: | Endoglucanase | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2010-08-19 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
2ZX6
 
 | | alpha-L-fucosidase complexed with inhibitor, F10-1C | | Descriptor: | 2-(1H-indol-3-yl)-N-{[(2R,3R,4R,5R,6S)-3,4,5-trihydroxy-6-methylpiperidin-2-yl]methyl}acetamide, Alpha-L-fucosidase, putative | | Authors: | Wu, H.-J, Ko, T.-P, Ho, C.-W, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2008-12-19 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural basis of alpha-fucosidase inhibition by iminocyclitols with K(i) values in the micro- to picomolar range.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
3AMH
 
 | | crystal structure of cellulase 12A from Thermotoga maritima | | Descriptor: | Endo-1,4-beta-glucanase | | Authors: | Cheng, Y.-S, Ko, T.-P, Liu, J.-R, Guo, R.-T. | | Deposit date: | 2010-08-20 | | Release date: | 2011-03-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure and substrate-binding mode of cellulase 12A from Thermotoga maritima
Proteins, 79, 2011
|
|
3AMR
 
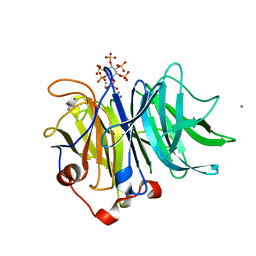 | | Crystal Structures of Bacillus subtilis Alkaline Phytase in Complex with Ca2+, Co2+, Ni2+, Mg2+ and myo-Inositol Hexasulfate | | Descriptor: | 3-phytase, CALCIUM ION, D-MYO-INOSITOL-HEXASULPHATE | | Authors: | Zeng, Y.F, Ko, T.P, Lai, H.L, Cheng, Y.S, Wu, T.H, Ma, Y, Yang, C.S, Cheng, K.J, Huang, C.H, Guo, R.T, Liu, J.R. | | Deposit date: | 2010-08-22 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structures of Bacillus alkaline phytase in complex with divalent metal ions and inositol hexasulfate
J.Mol.Biol., 409, 2011
|
|
3AMP
 
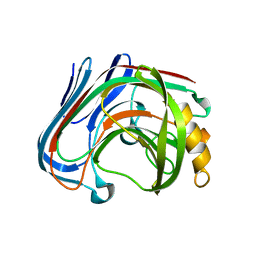 | | E134C-Cellotetraose complex of cellulase 12A from thermotoga maritima | | Descriptor: | Endo-1,4-beta-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Cheng, Y.-S, Ko, T.-P, Liu, J.-R, Guo, R.-T. | | Deposit date: | 2010-08-20 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure and substrate-binding mode of cellulase 12A from Thermotoga maritima
Proteins, 79, 2011
|
|
3AMC
 
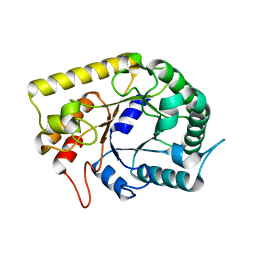 | | Crystal structures of Thermotoga maritima Cel5A, apo form and dimer/au | | Descriptor: | Endoglucanase | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2010-08-19 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3AMQ
 
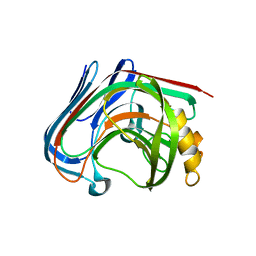 | | E134C-Cellobiose co-crystal of cellulase 12A from thermotoga maritima | | Descriptor: | Endo-1,4-beta-glucanase, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Cheng, Y.-S, Ko, T.-P, Liu, J.-R, Guo, R.-T. | | Deposit date: | 2010-08-20 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and substrate-binding mode of cellulase 12A from Thermotoga maritima
Proteins, 79, 2011
|
|
3AMG
 
 | | Crystal structures of Thermotoga maritima Cel5A in complex with Cellobiose substrate, mutant form | | Descriptor: | Endoglucanase, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2010-08-20 | | Release date: | 2011-08-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3AMS
 
 | | Crystal Structures of Bacillus subtilis Alkaline Phytase in Complex with Ca2+, Cd2+, Co2+, Ni2+, Mg2+ and myo-Inositol Hexasulfate | | Descriptor: | 3-phytase, CADMIUM ION, CALCIUM ION, ... | | Authors: | Zeng, Y.F, Ko, T.P, Lai, H.L, Cheng, Y.S, Wu, T.H, Ma, Y, Yang, C.S, Cheng, K.J, Huang, C.H, Guo, R.T, Liu, J.R. | | Deposit date: | 2010-08-22 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal structures of Bacillus alkaline phytase in complex with divalent metal ions and inositol hexasulfate
J.Mol.Biol., 409, 2011
|
|
3AOF
 
 | | Crystal structures of Thermotoga maritima Cel5A in complex with Mannotriose substrate | | Descriptor: | Endoglucanase, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2010-09-27 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.288 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3AMM
 
 | | Cellotetraose complex of cellulase 12A from thermotoga maritima | | Descriptor: | Endo-1,4-beta-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Cheng, Y.-S, Ko, T.-P, Liu, J.-R, Guo, R.-T. | | Deposit date: | 2010-08-20 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure and substrate-binding mode of cellulase 12A from Thermotoga maritima
Proteins, 79, 2011
|
|
3AMN
 
 | | E134C-Cellobiose complex of cellulase 12A from thermotoga maritima | | Descriptor: | Endo-1,4-beta-glucanase, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Cheng, Y.-S, Ko, T.-P, Liu, J.-R, Guo, R.-T. | | Deposit date: | 2010-08-20 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystal structure and substrate-binding mode of cellulase 12A from Thermotoga maritima
Proteins, 79, 2011
|
|
6LAA
 
 | | Crystal structure of full-length CYP116B46 from Tepidiphilus thermophilus | | Descriptor: | 1,2-ETHANEDIOL, CARBONATE ION, Cytochrome P450, ... | | Authors: | Zhang, L.L, Ko, T.P, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-11-12 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural insight into the electron transfer pathway of a self-sufficient P450 monooxygenase.
Nat Commun, 11, 2020
|
|
3AQ0
 
 | | Ligand-bound form of Arabidopsis medium/long-chain length prenyl pyrophosphate synthase (surface polar residue mutant) | | Descriptor: | 3-methylbut-3-enylsulfanyl(phosphonooxy)phosphinic acid, DI(HYDROXYETHYL)ETHER, FARNESYL DIPHOSPHATE, ... | | Authors: | Hsieh, F.-L, Chang, T.-H, Ko, T.-P, Wang, A.H.-J. | | Deposit date: | 2010-10-24 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure and mechanism of an Arabidopsis medium/long-chain-length prenyl pyrophosphate synthase
Plant Physiol., 155, 2011
|
|
3APZ
 
 | |
1M7J
 
 | | Crystal structure of D-aminoacylase defines a novel subset of amidohydrolases | | Descriptor: | ACETATE ION, D-aminoacylase, ZINC ION | | Authors: | Liaw, S.-H, Chen, S.-J, Ko, T.-P, Hsu, C.-S, Wang, A.H.-J, Tsai, Y.-C. | | Deposit date: | 2002-07-22 | | Release date: | 2003-02-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of D-Aminoacylase from Alcaligenes faecalis DA1. A NOVEL SUBSET OF AMIDOHYDROLASES AND INSIGHTS INTO THE ENZYME MECHANISM.
J.Biol.Chem., 278, 2003
|
|
4CXP
 
 | | Structure of bifunctional endonuclease (AtBFN2) from Arabidopsis thaliana in complex with sulfate | | Descriptor: | ENDONUCLEASE 2, PENTAETHYLENE GLYCOL, SULFATE ION, ... | | Authors: | Yu, T.-F, Maestre-Reyna, M, Ko, C.-Y, Ko, T.-P, Sun, Y.-J, Lin, T.-Y, Shaw, J.-F, Wang, A.H.-J. | | Deposit date: | 2014-04-08 | | Release date: | 2014-07-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Structural Insights of the Ssdna Binding Site in the Multifunctional Endonuclease Atbfn2 from Arabidopsis Thaliana.
Plos One, 9, 2014
|
|
4CWM
 
 | | High-glycosylation crystal structure of the bifunctional endonuclease (AtBFN2) from Arabidopsis thaliana | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ENDONUCLEASE 2, ZINC ION, ... | | Authors: | Yu, T.-F, Maestre-Reyna, M, Ko, C.-Y, Ko, T.-P, Sun, Y.-J, Lin, T.-Y, Shaw, J.-F, Wang, A.H.-J. | | Deposit date: | 2014-04-03 | | Release date: | 2014-07-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural Insights of the Ssdna Binding Site in the Multifunctional Endonuclease Atbfn2 from Arabidopsis Thaliana.
Plos One, 9, 2014
|
|
4CXO
 
 | | bifunctional endonuclease in complex with ssDNA | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, ENDONUCLEASE 2, SULFATE ION, ... | | Authors: | Yu, T.-F, Maestre-Reyna, M, Ko, C.-Y, Ko, T.-P, Sun, Y.-J, Lin, T.-Y, Shaw, J.-F, Wang, A.H.-J. | | Deposit date: | 2014-04-08 | | Release date: | 2014-07-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural Insights of the Ssdna Binding Site in the Multifunctional Endonuclease Atbfn2 from Arabidopsis Thaliana.
Plos One, 9, 2014
|
|
