3WGG
 
 | | Crystal structure of RSP in complex with alpha-NAD+ | | Descriptor: | Redox-sensing transcriptional repressor rex, SULFATE ION, alpha-Diphosphopyridine nucleotide | | Authors: | Zheng, Y, Ko, T.-P, Guo, R.-T. | | Deposit date: | 2013-08-05 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding mode of the oxidized alpha-anomer of NAD(+) to RSP, a Rex-family repressor
Biochem.Biophys.Res.Commun., 456, 2015
|
|
3WNQ
 
 | | Crystal structure of (R)-carbonyl reductase H49A mutant from Candida Parapsilosis in complex with 2-hydroxyacetophenone | | Descriptor: | (R)-specific carbonyl reductase, 2-hydroxy-1-phenylethanone, ZINC ION | | Authors: | Wang, S.S, Nie, Y, Xu, Y, Zhang, R.Z, Huang, C.H, Chan, H.C, Guo, R.T, Ko, T.P, Xiao, R. | | Deposit date: | 2013-12-15 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Unconserved substrate-binding sites direct the stereoselectivity of medium-chain alcohol dehydrogenase
Chem.Commun.(Camb.), 50, 2014
|
|
3WG9
 
 | | Crystal structure of RSP, a Rex-family repressor | | Descriptor: | Redox-sensing transcriptional repressor rex, SULFATE ION | | Authors: | Zheng, Y, Ko, T.-P, Guo, R.-T. | | Deposit date: | 2013-08-03 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Distinct structural features of Rex-family repressors to sense redox levels in anaerobes and aerobes.
J.Struct.Biol., 188, 2014
|
|
4QNN
 
 | |
3X2F
 
 | | A Thermophilic S-Adenosylhomocysteine Hydrolase | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Adenosylhomocysteinase, NITRATE ION, ... | | Authors: | Zheng, Y, Ko, T.P, Huang, C.H. | | Deposit date: | 2014-12-21 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structures of S-adenosylhomocysteine hydrolase from the thermophilic bacterium Thermotoga maritima.
J.Struct.Biol., 190, 2015
|
|
7W1D
 
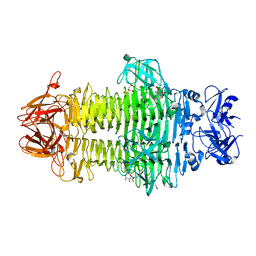 | | Crystal structure of Klebsiella pneumoniae K1 capsule-specific polysaccharide lyase in a C2 crystal form | | Descriptor: | CARBONATE ION, CITRIC ACID, K1 LYASE | | Authors: | Tu, I.F, Ko, T.P, Huang, K.F, Wu, S.H. | | Deposit date: | 2021-11-19 | | Release date: | 2022-05-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structural and biological insights into Klebsiella pneumoniae surface polysaccharide degradation by a bacteriophage K1 lyase: implications for clinical use.
J.Biomed.Sci., 29, 2022
|
|
3LBC
 
 | |
8HAD
 
 | |
8HAC
 
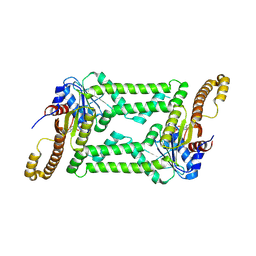 | |
7D4U
 
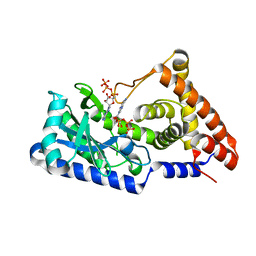 | | ATP complex with double mutant cyclic trinucleotide synthase CdnD | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cyclic AMP-AMP-GMP synthase | | Authors: | Yang, C.-S, Hou, M.-H, Tsai, C.-L, Wang, Y.-C, Ko, T.-P, Chen, Y. | | Deposit date: | 2020-09-24 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and functional implication of a bacterial cyclic AMP-AMP-GMP synthetase.
Nucleic Acids Res., 49, 2021
|
|
7D48
 
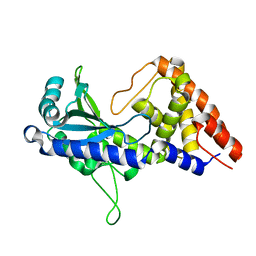 | | apo-form cyclic trinucleotide synthase CdnD | | Descriptor: | Cyclic AMP-AMP-GMP synthase, SODIUM ION | | Authors: | Yang, C.-S, Hou, M.-H, Tsai, C.-L, Wang, Y.-C, Ko, T.-P, Chen, Y. | | Deposit date: | 2020-09-23 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure and functional implication of a bacterial cyclic AMP-AMP-GMP synthetase.
Nucleic Acids Res., 49, 2021
|
|
7D4S
 
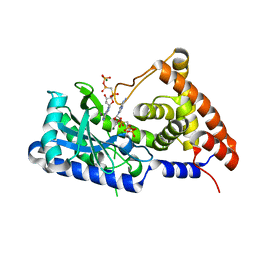 | | apo-form cyclic trinucleotide synthase CdnD | | Descriptor: | Cyclic AMP-AMP-GMP synthase, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION | | Authors: | Yang, C.-S, Hou, M.-H, Tsai, C.-L, Wang, Y.-C, Ko, T.-P, Chen, Y. | | Deposit date: | 2020-09-24 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure and functional implication of a bacterial cyclic AMP-AMP-GMP synthetase.
Nucleic Acids Res., 49, 2021
|
|
7D4J
 
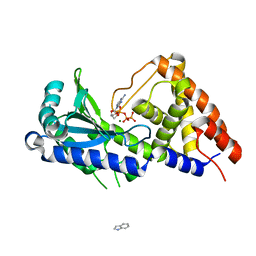 | | ddATP complex of cyclic trinucleotide synthase CdnD | | Descriptor: | 2',3'-dideoxyadenosine triphosphate, Cyclic AMP-AMP-GMP synthase, MAGNESIUM ION | | Authors: | Yang, C.-S, Hou, M.-H, Tsai, C.-L, Wang, Y.-C, Ko, T.-P, Chen, Y. | | Deposit date: | 2020-09-24 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure and functional implication of a bacterial cyclic AMP-AMP-GMP synthetase.
Nucleic Acids Res., 49, 2021
|
|
1V7U
 
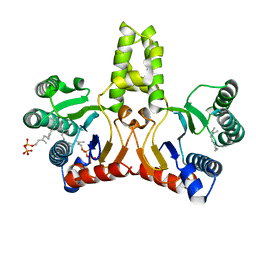 | | Crystal structure of Undecaprenyl Pyrophosphate Synthase with farnesyl pyrophosphate | | Descriptor: | FARNESYL DIPHOSPHATE, Undecaprenyl pyrophosphate synthetase | | Authors: | Chang, S.-Y, Ko, T.-P, Chen, A.P.-C, Wang, A.H.-J, Liang, P.-H. | | Deposit date: | 2003-12-24 | | Release date: | 2004-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Substrate binding mode and reaction mechanism of undecaprenyl pyrophosphate synthase deduced from crystallographic studies
Protein Sci., 13, 2004
|
|
6INR
 
 | | The crystal structure of phytoplasmal effector causing phyllody symptoms 1 (PHYL1) | | Descriptor: | CADMIUM ION, Putative effector, AYWB SAP54-like protein | | Authors: | Liao, Y.T, Lin, S.S, Ko, T.P, Wang, H.C. | | Deposit date: | 2018-10-26 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural insights into the interaction between phytoplasmal effector causing phyllody 1 and MADS transcription factors.
Plant J., 100, 2019
|
|
6JX7
 
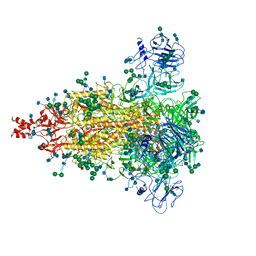 | | Cryo-EM structure of spike protein of feline infectious peritonitis virus strain UU4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Feline Infectious Peritonitis Virus Spike Protein, ... | | Authors: | Hsu, S.T.D, Yang, T.J, Ko, T.P, Draczkowski, P. | | Deposit date: | 2019-04-22 | | Release date: | 2020-01-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Cryo-EM analysis of a feline coronavirus spike protein reveals a unique structure and camouflaging glycans.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3PHL
 
 | | The apo-form UDP-glucose 6-dehydrogenase | | Descriptor: | UDP-glucose 6-dehydrogenase | | Authors: | Chen, Y.Y, Ko, T.P, Lin, C.H, Chen, W.H, Wang, A.H.J. | | Deposit date: | 2010-11-04 | | Release date: | 2011-09-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational change upon product binding to Klebsiella pneumoniae UDP-glucose dehydrogenase: a possible inhibition mechanism for the key enzyme in polymyxin resistance.
J.Struct.Biol., 175, 2011
|
|
3WL6
 
 | | Crystal Structure of pOPH Native | | Descriptor: | CITRIC ACID, Oxidized polyvinyl alcohol hydrolase | | Authors: | Yang, Y, Ko, T.P, Li, J.H, Liu, L, Huang, C.H, Chan, H.C, Ren, F.F, Jia, D.X, Wang, A.H.-J, Guo, R.T, Chen, J, Du, G.C. | | Deposit date: | 2013-11-08 | | Release date: | 2014-09-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into enzymatic degradation of oxidized polyvinyl alcohol
Chembiochem, 15, 2014
|
|
8J8Y
 
 | |
3WUF
 
 | | The mutant crystal structure of b-1,4-Xylanase (XynAS9_V43P/G44E) from Streptomyces sp. 9 | | Descriptor: | Endo-1,4-beta-xylanase A, ZINC ION | | Authors: | Chen, C.C, Han, X, Lv, P, Ko, T.P, Peng, W, Huang, C.H, Zheng, Y, Gao, J, Yang, Y.Y, Guo, R.T. | | Deposit date: | 2014-04-23 | | Release date: | 2014-10-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural perspectives of an engineered beta-1,4-xylanase with enhanced thermostability.
J.Biotechnol., 189C, 2014
|
|
3WE9
 
 | | The crystal structure of YisP from Bacillus subtilis subsp. subtilis strain 168 | | Descriptor: | Putative phytoene/squalene synthase YisP, TRIETHYLENE GLYCOL | | Authors: | Hu, Y, Huang, C.H, Chan, H.C, Ko, T.P, Feng, X, Oldfield, E, Guo, R.T. | | Deposit date: | 2013-07-02 | | Release date: | 2014-07-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of Bacillus subtilis YisP in complex with a PEG fragment
To be Published
|
|
3WWD
 
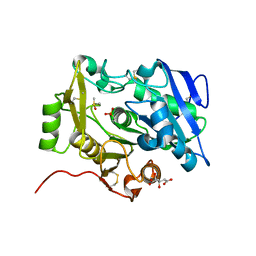 | | The complex of pOPH_S172C with DMSO | | Descriptor: | CITRIC ACID, DIMETHYL SULFOXIDE, Oxidized polyvinyl alcohol hydrolase | | Authors: | Yang, Y, Ko, T.P, Li, J.H, Liu, L, Huang, C.H, Chen, J, Guo, R.T, Du, G.C. | | Deposit date: | 2014-06-17 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Roles of tryptophan residue and disulfide bond in the variable lid region of oxidized polyvinyl alcohol hydrolase
Biochem.Biophys.Res.Commun., 452, 2014
|
|
3WJN
 
 | | Crystal structure of Octaprenyl Pyrophosphate synthase from Escherichia coli with farnesyl S-thiol-pyrophosphate (FSPP) | | Descriptor: | Octaprenyl diphosphate synthase, S-[(2E,6E)-3,7,11-TRIMETHYLDODECA-2,6,10-TRIENYL] TRIHYDROGEN THIODIPHOSPHATE | | Authors: | Han, X, Chen, C.C, Kuo, C.J, Huang, C.H, Zheng, Y, Ko, T.P, Zhu, Z, Feng, X, Oldfield, E, Liang, P.H, Guo, R.T, Ma, Y.H. | | Deposit date: | 2013-10-12 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of ligand-bound octaprenyl pyrophosphate synthase from Escherichia coli reveal the catalytic and chain-length determining mechanisms.
Proteins, 83, 2015
|
|
3WVJ
 
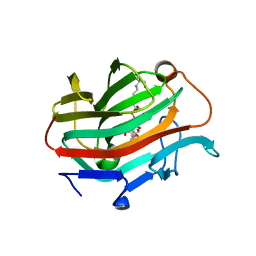 | | The crystal structure of native glycosidic hydrolase | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucanase | | Authors: | Chen, C.C, Huang, J.W, Zhao, P, Ko, T.P, Huang, C.H, Chan, H.C, Huang, Z, Liu, W, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-05-22 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analyses and yeast production of the beta-1,3-1,4-glucanase catalytic module encoded by the licB gene of Clostridium thermocellum.
Enzyme.Microb.Technol., 71, 2015
|
|
3WWC
 
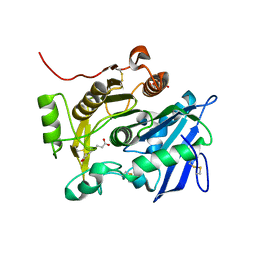 | | The complex of pOPH_S172A of pNPB | | Descriptor: | CITRIC ACID, Oxidized polyvinyl alcohol hydrolase, butanoic acid | | Authors: | Yang, Y, Ko, T.P, Li, J.H, Liu, L, Huang, C.H, Chen, J, Guo, R.T, Du, G.C. | | Deposit date: | 2014-06-17 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Roles of tryptophan residue and disulfide bond in the variable lid region of oxidized polyvinyl alcohol hydrolase
Biochem.Biophys.Res.Commun., 452, 2014
|
|
