4V8P
 
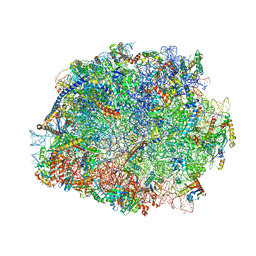 | | T.thermophila 60S ribosomal subunit in complex with initiation factor 6. | | Descriptor: | 26S RRNA, 5.8S RRNA, 5S RRNA, ... | | Authors: | Klinge, S, Voigts-Hoffmann, F, Leibundgut, M, Arpagaus, S, Ban, N. | | Deposit date: | 2011-09-14 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Crystal Structure of the Eukaryotic 60S Ribosomal Subunit in Complex with Initiation Factor 6.
Science, 334, 2011
|
|
5WLC
 
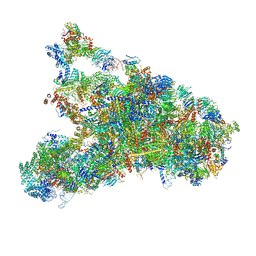 | | The complete structure of the small subunit processome | | Descriptor: | 18S pre-rRNA, 5' ETS, Bms1, ... | | Authors: | Barandun, J, Chaker-Margot, M, Hunziker, M, Klinge, S. | | Deposit date: | 2017-07-26 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The complete structure of the small-subunit processome.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5TZS
 
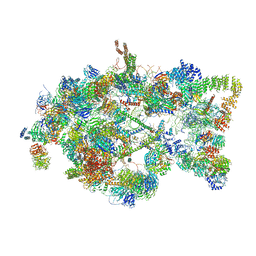 | | Architecture of the yeast small subunit processome | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S ribosomal RNA, 3' domain-associated, ... | | Authors: | Chaker-Margot, M, Barandun, J, Hunziker, M, Klinge, S. | | Deposit date: | 2016-11-22 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Architecture of the yeast small subunit processome.
Science, 355, 2017
|
|
6ND4
 
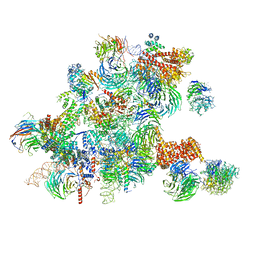 | | Conformational switches control early maturation of the eukaryotic small ribosomal subunit | | Descriptor: | 18S rRNA 5' domain start, 5'ETS rRNA, Bud21, ... | | Authors: | Hunziker, M, Barandun, J, Klinge, S. | | Deposit date: | 2018-12-13 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Conformational switches control early maturation of the eukaryotic small ribosomal subunit.
Elife, 8, 2019
|
|
4BSZ
 
 | |
4B08
 
 | | Yeast DNA polymerase alpha, Selenomethionine protein | | Descriptor: | DNA POLYMERASE ALPHA CATALYTIC SUBUNIT A | | Authors: | Perera, R.L, Torella, R, Klinge, S, Kilkenny, M.L, Maman, J.D, Pellegrini, L. | | Deposit date: | 2012-06-29 | | Release date: | 2013-02-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Mechanism for Priming DNA Synthesis by Yeast DNA Polymerase Alpha
Elife, 2, 2013
|
|
6RM3
 
 | | Evolutionary compaction and adaptation visualized by the structure of the dormant microsporidian ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 5S rRNA, ... | | Authors: | Barandun, J, Hunziker, M, Vossbrinck, C.R, Klinge, S. | | Deposit date: | 2019-05-05 | | Release date: | 2019-07-10 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Evolutionary compaction and adaptation visualized by the structure of the dormant microsporidian ribosome.
Nat Microbiol, 4, 2019
|
|
4FXD
 
 | | Crystal structure of yeast DNA polymerase alpha bound to DNA/RNA | | Descriptor: | DNA (5'-D(*TP*TP*TP*TP*CP*GP*CP*TP*GP*CP*CP*CP*GP*CP*CP*T)-3'), DNA polymerase alpha catalytic subunit A, RNA (5'-R(*AP*GP*GP*CP*GP*GP*GP*CP*AP*G)-3') | | Authors: | Perera, R.L, Torella, R, Klinge, S, Kilkenny, M.L, Maman, J.D, Pellegrini, L. | | Deposit date: | 2012-07-03 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanism for priming DNA synthesis by yeast DNA Polymerase alpha
eLife, 2, 2013
|
|
6C0F
 
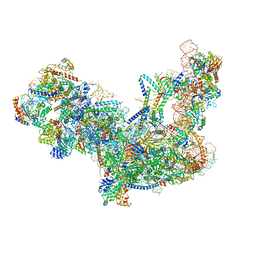 | | Yeast nucleolar pre-60S ribosomal subunit (state 2) | | Descriptor: | 5.8S rRNA, 60S ribosomal protein L13-A, 60S ribosomal protein L14-A, ... | | Authors: | Sanghai, Z.A, Miller, L, Barandun, J, Hunziker, M, Chaker-Margot, M, Klinge, S. | | Deposit date: | 2017-12-29 | | Release date: | 2018-03-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Modular assembly of the nucleolar pre-60S ribosomal subunit.
Nature, 556, 2018
|
|
6CB1
 
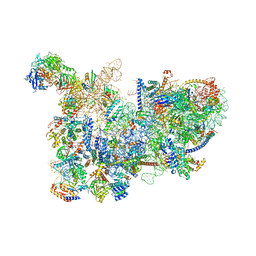 | | Yeast nucleolar pre-60S ribosomal subunit (state 3) | | Descriptor: | 35S pre-ribosomal RNA miscRNA, 5.8S rRNA, 60S ribosomal protein L13-A, ... | | Authors: | Sanghai, Z.A, Miller, L, Barandun, J, Hunziker, M, Chaker-Margot, M, Klinge, S. | | Deposit date: | 2018-02-01 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Modular assembly of the nucleolar pre-60S ribosomal subunit.
Nature, 556, 2018
|
|
7MQA
 
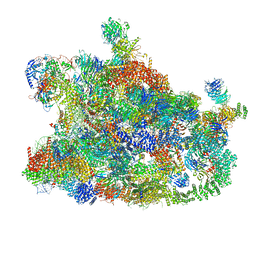 | | Cryo-EM structure of the human SSU processome, state post-A1 | | Descriptor: | 18S rRNA, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Vanden Broeck, A, Singh, S, Klinge, S. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Nucleolar maturation of the human small subunit processome.
Science, 373, 2021
|
|
7MQ8
 
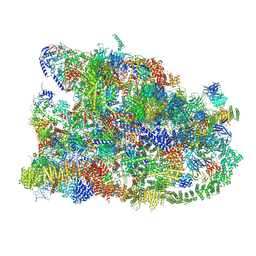 | | Cryo-EM structure of the human SSU processome, state pre-A1 | | Descriptor: | 18S rRNA, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Vanden Broeck, A, Singh, S, Klinge, S. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-22 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Nucleolar maturation of the human small subunit processome.
Science, 373, 2021
|
|
7MQJ
 
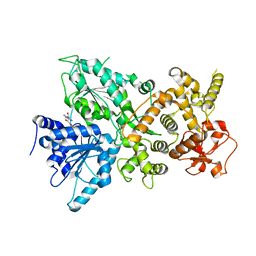 | | Dhr1 Helicase Core | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Probable ATP-dependent RNA helicase DHR1 | | Authors: | Miller, L, Chaker-Margot, M, Klinge, S. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Nucleolar maturation of the human small subunit processome.
Science, 373, 2021
|
|
7MQ9
 
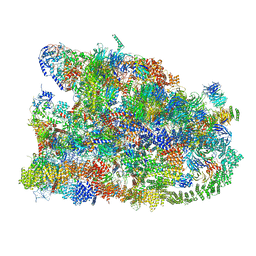 | | Cryo-EM structure of the human SSU processome, state pre-A1* | | Descriptor: | 18S rRNA, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Vanden Broeck, A, Singh, S, Klinge, S. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Nucleolar maturation of the human small subunit processome.
Science, 373, 2021
|
|
8CSR
 
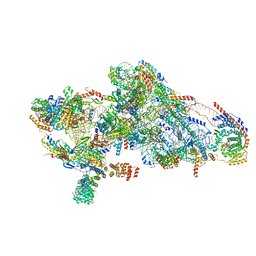 | | Human mitochondrial small subunit assembly intermediate (State C) | | Descriptor: | 12S mitochondrial rRNA, 28S ribosomal protein S10, mitochondrial, ... | | Authors: | Harper, N.J, Burnside, C, Klinge, S. | | Deposit date: | 2022-05-13 | | Release date: | 2022-12-14 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Principles of mitoribosomal small subunit assembly in eukaryotes.
Nature, 614, 2023
|
|
8CSS
 
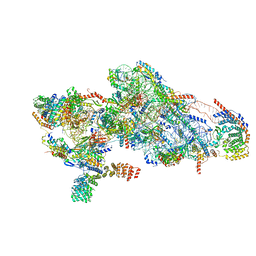 | | Human mitochondrial small subunit assembly intermediate (State D) | | Descriptor: | 12S mitochondrial rRNA, 28S ribosomal protein S10, mitochondrial, ... | | Authors: | Harper, N.J, Burnside, C, Klinge, S. | | Deposit date: | 2022-05-13 | | Release date: | 2022-12-14 | | Last modified: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Principles of mitoribosomal small subunit assembly in eukaryotes.
Nature, 614, 2023
|
|
8D8L
 
 | | Yeast mitochondrial small subunit assembly intermediate (State 3) | | Descriptor: | 15S ribosomal RNA, 3-hydroxyisobutyryl-CoA hydrolase, mitochondrial, ... | | Authors: | Burnside, C, Harper, N, Klinge, S. | | Deposit date: | 2022-06-08 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Principles of mitoribosomal small subunit assembly in eukaryotes.
Nature, 614, 2023
|
|
8CSP
 
 | | Human mitochondrial small subunit assembly intermediate (State A) | | Descriptor: | 12S mitochondrial rRNA, 28S ribosomal protein S10, mitochondrial, ... | | Authors: | Harper, N.J, Burnside, C, Klinge, S. | | Deposit date: | 2022-05-13 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Principles of mitoribosomal small subunit assembly in eukaryotes.
Nature, 614, 2023
|
|
8CSQ
 
 | | Human mitochondrial small subunit assembly intermediate (State B) | | Descriptor: | 12S mitochondrial rRNA, 28S ribosomal protein S10, mitochondrial, ... | | Authors: | Harper, N.J, Burnside, C, Klinge, S. | | Deposit date: | 2022-05-13 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Principles of mitoribosomal small subunit assembly in eukaryotes.
Nature, 614, 2023
|
|
8CSU
 
 | | Human mitochondrial small subunit assembly intermediate (State C*) | | Descriptor: | 12S mitochondrial rRNA, 28S ribosomal protein S10, mitochondrial, ... | | Authors: | Harper, N.J, Burnside, C, Klinge, S. | | Deposit date: | 2022-05-13 | | Release date: | 2022-12-14 | | Last modified: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Principles of mitoribosomal small subunit assembly in eukaryotes.
Nature, 614, 2023
|
|
8CST
 
 | | Human mitochondrial small subunit assembly intermediate (State E) | | Descriptor: | 12S mitochondrial rRNA, 28S ribosomal protein S10, mitochondrial, ... | | Authors: | Harper, N.J, Burnside, C, Klinge, S. | | Deposit date: | 2022-05-13 | | Release date: | 2022-12-14 | | Last modified: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Principles of mitoribosomal small subunit assembly in eukaryotes.
Nature, 614, 2023
|
|
8D8K
 
 | | Yeast mitochondrial small subunit assembly intermediate (State 2) | | Descriptor: | 15S ribosomal RNA, 37S ribosomal protein MRP1, mitochondrial, ... | | Authors: | Burnside, C, Harper, N.J, Klinge, S. | | Deposit date: | 2022-06-08 | | Release date: | 2022-12-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Principles of mitoribosomal small subunit assembly in eukaryotes.
Nature, 614, 2023
|
|
8D8J
 
 | | Yeast mitochondrial small subunit assembly intermediate (State 1) | | Descriptor: | 15S ribosomal RNA, 37S ribosomal protein MRP13, mitochondrial, ... | | Authors: | Burnside, C, Harper, N, Klinge, S. | | Deposit date: | 2022-06-08 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Principles of mitoribosomal small subunit assembly in eukaryotes.
Nature, 614, 2023
|
|
8FLF
 
 | |
8FKU
 
 | |
