3RCN
 
 | | Crystal Structure of Beta-N-Acetylhexosaminidase from Arthrobacter aurescens | | Descriptor: | ACETIC ACID, Beta-N-acetylhexosaminidase, GLYCEROL, ... | | Authors: | Kim, Y, Chhor, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-31 | | Release date: | 2011-06-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.514 Å) | | Cite: | Crystal Structure of Beta-N-Acetylhexosaminidase from Arthrobacter aurescens
To be Published
|
|
4Q6J
 
 | | Crystal Structure of EAL domain Protein from Listeria monocytogenes EGD-e | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, GLYCEROL, ... | | Authors: | Kim, Y, Bigelow, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-22 | | Release date: | 2014-05-07 | | Method: | X-RAY DIFFRACTION (1.369 Å) | | Cite: | Crystal Structure of EAL domain Protein from Listeria monocytogenes EGD-e
To be Published
|
|
3ONQ
 
 | | Crystal Structure of Regulator of Polyketide Synthase Expression BAD_0249 from Bifidobacterium adolescentis | | Descriptor: | GLYCEROL, Regulator of polyketide synthase expression, SULFATE ION | | Authors: | Kim, Y, Wu, R, Tan, K, Morales, J, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-30 | | Release date: | 2010-09-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Crystal Structure of Regulator of Polyketide Synthase Expression BAD_0249 from Bifidobacterium adolescentis
To be Published
|
|
4Q62
 
 | | Crystal Structure of Leucine-rich repeat- and Coiled coil-containing Protein from Legionella pneumophila | | Descriptor: | 1,2-ETHANEDIOL, Leucine-rich repeat-and coiled coil-containing protein, SULFATE ION | | Authors: | Kim, Y, Hatzos-Skintges, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2014-04-20 | | Release date: | 2014-05-07 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Crystal Structure of Leucine-rich repeat- and Coiled coil-containing Protein from
Legionella pneumophila
To be Published
|
|
7F27
 
 | | Crystal structure of polyketide ketosynthase | | Descriptor: | 3-oxoacyl-(Acyl-carrier-protein) synthase | | Authors: | Kim, Y, Lee, W.C. | | Deposit date: | 2021-06-10 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.806 Å) | | Cite: | Structural basis of the complementary activity of two ketosynthases in aryl polyene biosynthesis.
Sci Rep, 11, 2021
|
|
4Q7A
 
 | | Crystal Structure of N-acetyl-ornithine/N-acetyl-lysine Deacetylase from Sphaerobacter thermophilus | | Descriptor: | CHLORIDE ION, GLYCEROL, N-acetyl-ornithine/N-acetyl-lysine deacetylase, ... | | Authors: | Kim, Y, Tesar, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-24 | | Release date: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (2.048 Å) | | Cite: | Crystal Structure of N-acetyl-ornithine/N-acetyl-lysine Deacetylase from
Sphaerobacter thermophilus
To be Published
|
|
3RE3
 
 | | Crystal Structure of 2-C-Methyl-D-Erythritol 2,4-Cyclodiphosphate Synthase from Francisella tularensis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, CHLORIDE ION, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-04-02 | | Release date: | 2011-04-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.645 Å) | | Cite: | Crystal Structure of 2-C-Methyl-D-Erythritol 2,4-Cyclodiphosphate Synthase from Francisella tularensis
To be Published
|
|
3P2L
 
 | | Crystal Structure of ATP-dependent Clp protease subunit P from Francisella tularensis | | Descriptor: | 1,2-ETHANEDIOL, ATP-dependent Clp protease proteolytic subunit, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Zhou, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-10-02 | | Release date: | 2010-10-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Crystal Structure of ATP-dependent Clp protease subunit P from Francisella tularensis
To be Published
|
|
3SBL
 
 | | Crystal Structure of New Delhi Metal-beta-lactamase-1 from Klebsiella pneumoniae | | Descriptor: | Beta-lactamase NDM-1, CITRIC ACID | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, J, Binkowski, T.A, Mire, J, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2011-06-05 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure of Apo- and Monometalated Forms of NDM-1 A Highly Potent Carbapenem-Hydrolyzing Metallo-beta-Lactamase
Plos One, 6, 2011
|
|
4R9O
 
 | | Crystal Structure of Putative Aldo/Keto Reductase from Salmonella enterica | | Descriptor: | Putative aldo/keto reductase | | Authors: | Kim, Y, Maltseva, N, Stam, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-09-05 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Crystal Structure of Putative Aldo/Keto Reductase from Salmonella enterica
To be Published
|
|
2PDO
 
 | | Crystal Structure of the Putative Acetyltransferase of GNAT Family from Shigella flexneri | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Acetyltransferase ypeA, ... | | Authors: | Kim, Y, Li, H, Holzle, D, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-04-01 | | Release date: | 2007-04-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Putative Acetyltransferase of GNAT Family from Shigella flexneri
To be Published
|
|
7CAX
 
 | | Crystal structure of bacterial reductase | | Descriptor: | 1,2-ETHANEDIOL, 3-oxoacyl-ACP reductase FabG, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Kim, Y, Lee, W.C. | | Deposit date: | 2020-06-10 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.846 Å) | | Cite: | Crystal structure of bacterial reductase
To be published
|
|
4NMU
 
 | | Crystal Structure of Thiol-disulfide Oxidoreductase from Bacillus str. 'Ames Ancestor' | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Zhou, M, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-15 | | Release date: | 2013-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure of Thiol-disulfide Oxidoreductase from Bacillus str. 'Ames Ancestor'
To be Published
|
|
4MY1
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Bacillus anthracis str. Ames complexed with P68 | | Descriptor: | 1-(4-bromophenyl)-3-(2-{3-[(1E)-N-hydroxyethanimidoyl]phenyl}propan-2-yl)urea, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-26 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5997 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Bacillus anthracis str. Ames complexed with P68
To be Published
|
|
4PIB
 
 | | Crystal Structure of Uncharacterized Conserved Protein PixA from Burkholderia thailandensis | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kim, Y, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-05-08 | | Release date: | 2014-07-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Uncharacterized Conserved Protein PixA from Burkholderia thailandensis
To Be Published
|
|
4MY9
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor C91 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, MALONATE ION, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-27 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5893 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor C91
To be Published
|
|
3R7T
 
 | | Crystal Structure of Adenylosuccinate Synthetase from Campylobacter jejuni | | Descriptor: | Adenylosuccinate synthetase, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Kim, Y, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-23 | | Release date: | 2011-04-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Adenylosuccinate Synthetase from
Campylobacter jejuni
To be Published
|
|
3TSD
 
 | | Crystal Structure of Inosine-5'-monophosphate Dehydrogenase from Bacillus anthracis str. Ames complexed with XMP | | Descriptor: | D(-)-TARTARIC ACID, Inosine-5'-monophosphate dehydrogenase, SULFATE ION, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-13 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.653 Å) | | Cite: | Bacillus anthracis inosine 5'-monophosphate dehydrogenase in action: the first bacterial series of structures of phosphate ion-, substrate-, and product-bound complexes.
Biochemistry, 51, 2012
|
|
3TSB
 
 | | Crystal Structure of Inosine-5'-monophosphate Dehydrogenase from Bacillus anthracis str. Ames | | Descriptor: | Inosine-5'-monophosphate dehydrogenase, PHOSPHATE ION | | Authors: | Kim, Y, Makowska-Grzyska, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-12 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Bacillus anthracis inosine 5'-monophosphate dehydrogenase in action: the first bacterial series of structures of phosphate ion-, substrate-, and product-bound complexes.
Biochemistry, 51, 2012
|
|
4R7O
 
 | | Crystal Structure of Putative Glycerophosphoryl Diester Phosphodiesterasefrom Bacillus anthraci | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kim, Y, Zhou, M, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-28 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.534 Å) | | Cite: | Crystal Structure of Putative Glycerophosphoryl Diester Phosphodiesterasefrom Bacillus anthraci
To be Published
|
|
4MY8
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor Q21 | | Descriptor: | (2S)-2-(naphthalen-1-yloxy)-N-[2-(pyridin-4-yl)-1,3-benzoxazol-5-yl]propanamide, 1,2-ETHANEDIOL, ACETIC ACID, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Gorla, S.K, Kavitha, M, Cuny, G, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-27 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2924 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor Q21
To be Published, 2013
|
|
4MZ8
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with an Internal Deletion of CBS Domain from Campylobacter jejuni complexed with inhibitor compound C91 | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-29 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5004 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Campylobacter jejuni complexed with inhibitor compound C91
To be Published
|
|
3USB
 
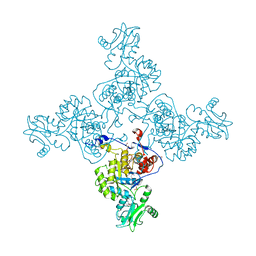 | | Crystal Structure of Bacillus anthracis Inosine Monophosphate Dehydrogenase in the complex with IMP | | Descriptor: | CHLORIDE ION, GLYCEROL, INOSINIC ACID, ... | | Authors: | Kim, Y, Zhang, R, Wu, R, Gu, M, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-11-23 | | Release date: | 2011-12-07 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Bacillus anthracis inosine 5'-monophosphate dehydrogenase in action: the first bacterial series of structures of phosphate ion-, substrate-, and product-bound complexes.
Biochemistry, 51, 2012
|
|
4MY0
 
 | | Crystal Structure of GCN5-related N-acetyltransferase from Kribbella flavida | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETYL COENZYME *A, GCN5-related N-acetyltransferase, ... | | Authors: | Kim, Y, Mack, J, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-26 | | Release date: | 2013-11-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Crystal Structure of GCN5-related N-acetyltransferase from Kribbella flavida
To be Published
|
|
7EWP
 
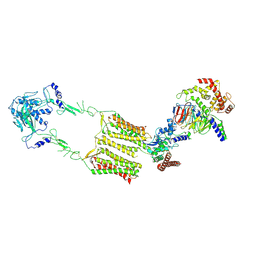 | | Cryo-EM structure of human GPR158 in complex with RGS7-Gbeta5 in a 2:1:1 ratio | | Descriptor: | Guanine nucleotide-binding protein subunit beta-5, Probable G-protein coupled receptor 158, Regulator of G-protein signaling 7 | | Authors: | Kim, Y, Jeong, E, Jeong, J, Cho, Y. | | Deposit date: | 2021-05-25 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure of the class C orphan GPCR GPR158 in complex with RGS7-G beta 5.
Nat Commun, 12, 2021
|
|
