7PA2
 
 | | PARK7 with inhibitor 8RK64 | | Descriptor: | (3~{S})-~{N}-[5-[2-[(azanylidene-$l^{4}-azanylidene)amino]ethanoyl]-6,7-dihydro-4~{H}-[1,3]thiazolo[5,4-c]pyridin-2-yl]-1-(iminomethyl)pyrrolidine-3-carboxamide, Parkinson disease protein 7 | | Authors: | Kim, R.Q, Jia, Y, Sapmaz, A, Geurink, P.P. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Chemical Toolkit for PARK7: Potent, Selective, and High-Throughput.
J.Med.Chem., 65, 2022
|
|
7PA3
 
 | | PARK7 with covalent inhibitor JYQ-88 | | Descriptor: | (3~{S})-~{N}-[5-[2-[(azanylidene-$l^{4}-azanylidene)amino]ethanoyl]-6,7-dihydro-4~{H}-[1,3]thiazolo[5,4-c]pyridin-2-yl]-1-(iminomethyl)pyrrolidine-3-carboxamide, Parkinson disease protein 7 | | Authors: | Kim, R.Q, Jia, Y, Sapmaz, A, Geurink, P.P. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Chemical Toolkit for PARK7: Potent, Selective, and High-Throughput.
J.Med.Chem., 65, 2022
|
|
4BMZ
 
 | | Structure of futalosine hydrolase mutant of Helicobacter pylori strain 26695 | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, MTA/SAH NUCLEOSIDASE | | Authors: | Kim, R.Q, Offen, W.A, Stubbs, K.A, Davies, G.J. | | Deposit date: | 2013-05-12 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Enzymology of Helicobacter Pylori Methylthioadenosine Nucleosidase in the Futalosine Pathway
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4BN0
 
 | | Structure of futalosine hydrolase mutant of Helicobacter pylori strain 26695 | | Descriptor: | MTA/SAH NUCLEOSIDASE | | Authors: | Kim, R.Q, Offen, W.A, Stubbs, K.A, Davies, G.J. | | Deposit date: | 2013-05-12 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural Enzymology of Helicobacter Pylori Methylthioadenosine Nucleosidase in the Futalosine Pathway
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4BMY
 
 | | Structure of futalosine hydrolase mutant of Helicobacter pylori strain 26695 | | Descriptor: | MTA/SAH NUCLEOSIDASE, SULFATE ION | | Authors: | Kim, R.Q, Offen, W.A, Stubbs, K.A, Davies, G.J. | | Deposit date: | 2013-05-12 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Enzymology of Helicobacter Pylori Methylthioadenosine Nucleosidase in the Futalosine Pathway
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4BMX
 
 | | Native structure of futalosine hydrolase of Helicobacter pylori strain 26695 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENINE, MTA/SAH NUCLEOSIDASE | | Authors: | Kim, R.Q, Offen, W.A, Stubbs, K.A, Davies, G.J. | | Deposit date: | 2013-05-12 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Enzymology of Helicobacter Pylori Methylthioadenosine Nucleosidase in the Futalosine Pathway
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5FWI
 
 | |
6T9D
 
 | | Crystal structure of a bispecific DutaFab in complex with human VEGF121 | | Descriptor: | VP mat DutaFab VH chain, VP mat DutaFab VL chain, Vascular endothelial growth factor A | | Authors: | Kimbung, R, Logan, D.T, Beckmann, R, Jensen, K, Speck, J, Fenn, S, Kettenberger, H. | | Deposit date: | 2019-10-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.905 Å) | | Cite: | DutaFabs are engineered therapeutic Fab fragments that can bind two targets simultaneously.
Nat Commun, 12, 2021
|
|
6T9E
 
 | | Crystal structure of a bispecific DutaFab in complex with human PDGF | | Descriptor: | DutaFab mat VH chain, DutaFab mat VL chain, Platelet-derived growth factor subunit B | | Authors: | Kimbung, R, Logan, D.T, Beckmann, R, Jensen, K, Speck, J, Fenn, S, Kettenberger, H. | | Deposit date: | 2019-10-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.989 Å) | | Cite: | DutaFabs are engineered therapeutic Fab fragments that can bind two targets simultaneously.
Nat Commun, 12, 2021
|
|
1KJY
 
 | | Crystal Structure of Human G[alpha]i1 Bound to the GoLoco Motif of RGS14 | | Descriptor: | CESIUM ION, GUANINE NUCLEOTIDE-BINDING PROTEIN G(I), ALPHA-1 SUBUNIT, ... | | Authors: | Kimple, R.J, Kimple, M.E, Betts, L, Sondek, J, Siderovski, D.P. | | Deposit date: | 2001-12-05 | | Release date: | 2002-05-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural determinants for GoLoco-induced inhibition of nucleotide release by Galpha subunits.
Nature, 416, 2002
|
|
6CDX
 
 | | High-resolution crystal structure of fluoropropylated cystine knot, binding to alpha-5 beta-6 integrin | | Descriptor: | cystine knot (fluoropropylated) | | Authors: | Kimura, R, Nix, J, Bongura, C, Chakraborti, S, Gambhir, S, Filipp, F.V. | | Deposit date: | 2018-02-09 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Evaluation of integrin alpha v beta6cystine knot PET tracers to detect cancer and idiopathic pulmonary fibrosis.
Nat Commun, 10, 2019
|
|
1MRZ
 
 | | Crystal structure of a flavin binding protein from Thermotoga Maritima, TM379 | | Descriptor: | CITRIC ACID, Riboflavin kinase/FMN adenylyltransferase | | Authors: | Wang, W, Kim, R, Jancarik, J, Yokota, H, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-09-19 | | Release date: | 2003-09-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a flavin-binding protein from Thermotoga Maritima
Proteins, 52, 2003
|
|
1EKE
 
 | | CRYSTAL STRUCTURE OF CLASS II RIBONUCLEASE H (RNASE HII) WITH MES LIGAND | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, RIBONUCLEASE HII | | Authors: | Lai, L.H, Yokota, H, Hung, L.W, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2000-03-07 | | Release date: | 2000-09-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of archaeal RNase HII: a homologue of human major RNase H
Structure, 8, 2000
|
|
1N0F
 
 | | CRYSTAL STRUCTURE OF A CELL DIVISION AND CELL WALL BIOSYNTHESIS PROTEIN UPF0040 FROM MYCOPLASMA PNEUMONIAE: INDICATION OF A NOVEL FOLD WITH A POSSIBLE NEW CONSERVED SEQUENCE MOTIF | | Descriptor: | Protein mraZ | | Authors: | Chen, S, Jancrick, J, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-10-13 | | Release date: | 2003-10-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a protein associated with cell division from Mycoplasma pneumoniae (GI: 13508053): a novel fold with a conserved sequence motif.
Proteins, 55, 2004
|
|
1N0E
 
 | | CRYSTAL STRUCTURE OF A CELL DIVISION AND CELL WALL BIOSYNTHESIS PROTEIN UPF0040 FROM MYCOPLASMA PNEUMONIAE: INDICATION OF A NOVEL FOLD WITH A POSSIBLE NEW CONSERVED SEQUENCE MOTIF | | Descriptor: | Protein mraZ | | Authors: | Chen, S, Jancrick, J, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-10-13 | | Release date: | 2003-10-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a protein associated with cell division from Mycoplasma pneumoniae (GI: 13508053): a novel fold with a conserved sequence motif.
Proteins, 55, 2004
|
|
1N0G
 
 | | Crystal Structure of A Cell Division and Cell Wall Biosynthesis Protein UPF0040 from Mycoplasma pneumoniae: Indication of A Novel Fold with A Possible New Conserved Sequence Motif | | Descriptor: | Protein mraZ | | Authors: | Chen, S, Jancarik, J, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-10-13 | | Release date: | 2003-10-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a protein associated with cell division from Mycoplasma pneumoniae (GI: 13508053): a novel fold with a conserved sequence motif.
Proteins, 55, 2004
|
|
4R3R
 
 | | Crystal structures of EGFR in complex with Mig6 | | Descriptor: | Epidermal growth factor receptor, peptide from ERBB receptor feedback inhibitor 1' | | Authors: | Park, E, Kim, N, Yi, Z, Cho, A, Kim, K, Ficarro, S.B, Park, A, Park, W.Y, Murray, B, Meyerson, M, Beroukim, R, Marto, J.A, Cho, J, Eck, M.J. | | Deposit date: | 2014-08-17 | | Release date: | 2015-08-12 | | Last modified: | 2015-09-16 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure and mechanism of activity-based inhibition of the EGF receptor by Mig6.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4R3P
 
 | | Crystal structures of EGFR in complex with Mig6 | | Descriptor: | Epidermal growth factor receptor, peptide from ERBB receptor feedback inhibitor 1 | | Authors: | Park, E, Kim, N, Yi, Z, Cho, A, Kim, K, Ficarro, S.B, Park, A, Park, W.Y, Murray, B, Meyerson, M, Beroukim, R, Marto, J.A, Cho, J, Eck, M.J. | | Deposit date: | 2014-08-17 | | Release date: | 2015-08-12 | | Last modified: | 2015-09-16 | | Method: | X-RAY DIFFRACTION (2.905 Å) | | Cite: | Structure and mechanism of activity-based inhibition of the EGF receptor by Mig6.
Nat.Struct.Mol.Biol., 22, 2015
|
|
1EIF
 
 | | EUKARYOTIC TRANSLATION INITIATION FACTOR 5A FROM METHANOCOCCUS JANNASCHII | | Descriptor: | EUKARYOTIC TRANSLATION INITIATION FACTOR 5A | | Authors: | Kim, K.K, Hung, L.W, Yokota, H, Kim, R, Kim, S.H. | | Deposit date: | 1998-07-29 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of eukaryotic translation initiation factor 5A from Methanococcus jannaschii at 1.8 A resolution.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1FBN
 
 | | CRYSTAL STRUCTURE OF A FIBRILLARIN HOMOLOGUE FROM METHANOCOCCUS JANNASCHII, A HYPERTHERMOPHILE, AT 1.6 A | | Descriptor: | MJ FIBRILLARIN HOMOLOGUE | | Authors: | Wang, H, Boisvert, D, Kim, K.K, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 1999-04-25 | | Release date: | 2000-04-26 | | Last modified: | 2014-11-26 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a fibrillarin homologue from Methanococcus jannaschii, a hyperthermophile, at 1.6 A resolution.
EMBO J., 19, 2000
|
|
2EIF
 
 | | Eukaryotic translation initiation factor 5A from Methanococcus jannaschii | | Descriptor: | PROTEIN (EUKARYOTIC TRANSLATION INITIATION FACTOR 5A) | | Authors: | Kim, K.K, Hung, L.W, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 1998-10-12 | | Release date: | 1999-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of eukaryotic translation initiation factor 5A from Methanococcus jannaschii at 1.8 A resolution.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1F5S
 
 | | CRYSTAL STRUCTURE OF PHOSPHOSERINE PHOSPHATASE FROM METHANOCOCCUS JANNASCHII | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, PHOSPHOSERINE PHOSPHATASE (PSP) | | Authors: | Wang, W, Kim, R, Jancarik, J, Yokota, H, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2000-06-15 | | Release date: | 2001-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of phosphoserine phosphatase from Methanococcus jannaschii, a hyperthermophile, at 1.8 A resolution.
Structure, 9, 2001
|
|
1G2I
 
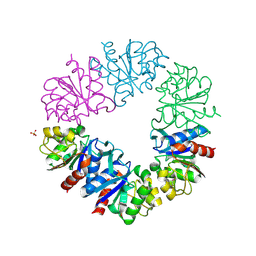 | | CRYSTAL STRUCTURE OF A NOVEL INTRACELLULAR PROTEASE FROM PYROCOCCUS HORIKOSHII AT 2 A RESOLUTION | | Descriptor: | PROTEASE I, SULFATE ION | | Authors: | Du, X, Choi, I.-G, Kim, R, Jancarik, J, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2000-10-19 | | Release date: | 2000-11-08 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an intracellular protease from Pyrococcus horikoshii at 2-A resolution.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1STZ
 
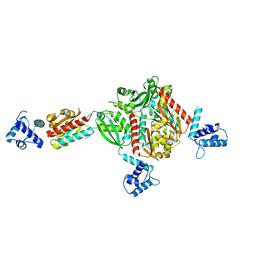 | | Crystal structure of a hypothetical protein at 2.2 A resolution | | Descriptor: | Heat-inducible transcription repressor hrcA homolog | | Authors: | Liu, J, Adams, P.D, Shin, D.-H, Huang, C, Yokota, H, Jancarik, J, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-03-25 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a heat-inducible transcriptional repressor HrcA from Thermotoga maritima: structural insight into DNA binding and dimerization.
J.Mol.Biol., 350, 2005
|
|
1Q8C
 
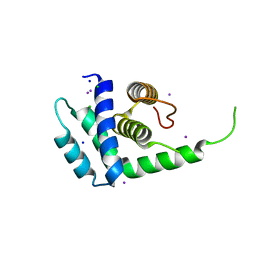 | | A conserved hypothetical protein from Mycoplasma genitalium shows structural homology to NusB proteins | | Descriptor: | CHLORIDE ION, Hypothetical protein MG027, IODIDE ION, ... | | Authors: | Liu, J, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2003-08-20 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A conserved hypothetical protein from Mycoplasma genitalium shows structural homology to nusb proteins
Proteins, 55, 2004
|
|
