7ER9
 
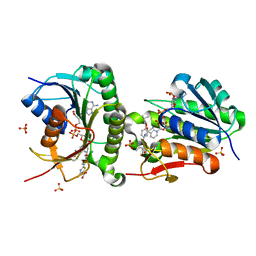 | | Crystal structure of human Biliverdin IX-beta reductase B with Febuxostat (TEI) | | 分子名称: | 2-(3-CYANO-4-ISOBUTOXY-PHENYL)-4-METHYL-5-THIAZOLE-CARBOXYLIC ACID, Flavin reductase (NADPH), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Griesinger, C, Lee, D, Ryu, K.S, Kim, M, Ha, J.H. | | 登録日 | 2021-05-06 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Repositioning Food and Drug Administration-Approved Drugs for Inhibiting Biliverdin IX beta Reductase B as a Novel Thrombocytopenia Therapeutic Target.
J.Med.Chem., 65, 2022
|
|
7ERA
 
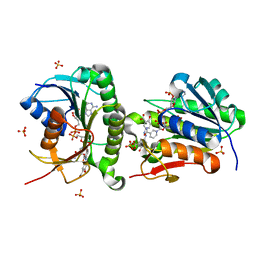 | | Crystal structure of human Biliverdin IX-beta reductase B with Olsalazine Sodium (OSS) | | 分子名称: | 5-[(E)-(3-carboxy-4-oxidanyl-phenyl)diazenyl]-2-oxidanyl-benzoic acid, Flavin reductase (NADPH), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Griesinger, C, Lee, D, Ryu, K.S, Kim, M, Ha, J.H. | | 登録日 | 2021-05-06 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Repositioning Food and Drug Administration-Approved Drugs for Inhibiting Biliverdin IX beta Reductase B as a Novel Thrombocytopenia Therapeutic Target.
J.Med.Chem., 65, 2022
|
|
7ER6
 
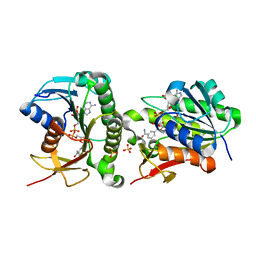 | | Crystal structure of human Biliverdin IX-beta reductase B | | 分子名称: | Flavin reductase (NADPH), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | 著者 | Griesinger, C, Lee, D, Ryu, K.S, Kim, M, Ha, J.H. | | 登録日 | 2021-05-06 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Repositioning Food and Drug Administration-Approved Drugs for Inhibiting Biliverdin IX beta Reductase B as a Novel Thrombocytopenia Therapeutic Target.
J.Med.Chem., 65, 2022
|
|
7ERE
 
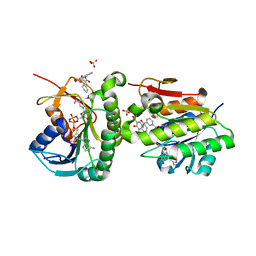 | | Crystal structure of human Biliverdin IX-beta reductase B with Pyrantel Pamoate (PPA) | | 分子名称: | 4-[(3-carboxy-2-oxidanyl-naphthalen-1-yl)methyl]-3-oxidanyl-naphthalene-2-carboxylic acid, Flavin reductase (NADPH), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Griesinger, C, Lee, D, Ryu, K.S, Kim, M, Ha, J.H. | | 登録日 | 2021-05-06 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Repositioning Food and Drug Administration-Approved Drugs for Inhibiting Biliverdin IX beta Reductase B as a Novel Thrombocytopenia Therapeutic Target.
J.Med.Chem., 65, 2022
|
|
7ERD
 
 | | Crystal structure of human Biliverdin IX-beta reductase B with Flunixin Meglumin (FMG) | | 分子名称: | 2-[[2-methyl-3-(trifluoromethyl)phenyl]amino]pyridine-3-carboxylic acid, Flavin reductase (NADPH), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Griesinger, C, Lee, D, Ryu, K.S, Kim, M, Ha, J.H. | | 登録日 | 2021-05-06 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Repositioning Food and Drug Administration-Approved Drugs for Inhibiting Biliverdin IX beta Reductase B as a Novel Thrombocytopenia Therapeutic Target.
J.Med.Chem., 65, 2022
|
|
1IU7
 
 | | HOLO FORM OF COPPER-CONTAINING AMINE OXIDASE FROM ARTHROBACTER GLOBIFORMIS | | 分子名称: | AMINE OXIDASE, COPPER (II) ION | | 著者 | Kishishita, S, Okajima, T, Kim, M, Yamaguchi, H, Hirota, S, Suzuki, S, Kuroda, S, Tanizawa, K, Mure, M. | | 登録日 | 2002-02-28 | | 公開日 | 2003-02-04 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Role of Copper Ion in Bacterial Copper Amine Oxidase: Spectroscopic and Crystallographic Studies of Metal-Substituted Enzymes
J.AM.CHEM.SOC., 125, 2003
|
|
1QA9
 
 | | Structure of a Heterophilic Adhesion Complex Between the Human CD2 and CD58(LFA-3) Counter-Receptors | | 分子名称: | HUMAN CD2 PROTEIN, HUMAN CD58 PROTEIN | | 著者 | Wang, J.-H, Smolyar, A, Tan, K, Liu, J.-H, Kim, M, Sun, Z.J, Wagner, G, Reinherz, E.L. | | 登録日 | 1999-04-13 | | 公開日 | 1999-04-29 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structure of a heterophilic adhesion complex between the human CD2 and CD58 (LFA-3) counterreceptors.
Cell(Cambridge,Mass.), 97, 1999
|
|
1IQX
 
 | | CRYSTAL STRUCTURE OF COBALT-SUBSTITUTED AMINE OXIDASE FROM ARTHROBACTER GLOBIFORMIS | | 分子名称: | CO(II)-SUBSTITUTED AMINE OXIDASE, COBALT (II) ION | | 著者 | Kishishita, S, Okajima, T, Mure, M, Kim, M, Yamaguchi, H, Hirota, S, Suzuki, S, Kuroda, S, Tanizawa, K. | | 登録日 | 2001-08-27 | | 公開日 | 2003-02-04 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Role of Copper Ion in Bacterial Copper Amine Oxidase: Spectroscopic and Crystallographic Studies of Metal-Substituted Enzymes
J.AM.CHEM.SOC., 125, 2003
|
|
1IQY
 
 | | CRYSTAL STRUCTURE OF NICKEL-SUBSTITUTED AMINE OXIDASE FROM ARTHROBACTER GLOBIFORMIS | | 分子名称: | AMINE OXIDASE, NICKEL (II) ION | | 著者 | Kishishita, S, Okajima, T, Mure, M, Kim, M, Yamaguchi, H, Hirota, S, Kuroda, S, Tanizawa, K. | | 登録日 | 2001-08-28 | | 公開日 | 2003-02-04 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Role of Copper Ion in Bacterial Copper Amine Oxidase: Spectroscopic and Crystallographic Studies of Metal-Substituted Enzymes
J.AM.CHEM.SOC., 125, 2003
|
|
6K6I
 
 | | The crystal structure of light-driven cyanobacterial chloride importer from Mastigocladopsis repens | | 分子名称: | CHLORIDE ION, Cyanobacterial chloride importer, OLEIC ACID, ... | | 著者 | Yun, J.H, Park, J.H, Jin, Z, Ohki, M, Wang, Y, Lupala, C.S, Kim, M, Liu, H, Park, S.Y, Lee, W. | | 登録日 | 2019-06-03 | | 公開日 | 2020-06-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The crystal structure of light-driven cyanobacterial chloride importer from Mastigocladopsis repens
To Be Published
|
|
6K6K
 
 | | The crystal structure of light-driven cyanobacterial chloride importer (N63A/P118A) Mastigocladopsis repens | | 分子名称: | CHLORIDE ION, Cyanobacterial chloride importer, OLEIC ACID, ... | | 著者 | Yun, J.H, Park, J.H, Jin, Z, Ohki, M, Wang, Y, Lupala, C.S, Kim, M, Liu, H, Park, S.Y, Lee, W. | | 登録日 | 2019-06-03 | | 公開日 | 2020-06-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.197 Å) | | 主引用文献 | The crystal structure of light-driven cyanobacterial chloride importer (N63A/P118A) Mastigocladopsis repens
To Be Published
|
|
7VYT
 
 | | Crystal structure of human TIGIT(23-129) in complex with the scFv fragment of anti-TIGIT antibody MG1131 | | 分子名称: | CITRATE ANION, MG1131 heavy chain variable region, MG1131 light chain variable region, ... | | 著者 | Jeong, B.-S, Nam, H, Kim, M, Oh, B.-H. | | 登録日 | 2021-11-15 | | 公開日 | 2022-03-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | Structural and functional characterization of a monoclonal antibody blocking TIGIT.
Mabs, 14, 2022
|
|
6K6J
 
 | | The crystal structure of light-driven cyanobacterial chloride importer from Mastigocladopsis repens with Bromide ion | | 分子名称: | BROMIDE ION, Cyanobacterial chloride importer, OLEIC ACID, ... | | 著者 | Yun, J.H, Park, J.H, Jin, Z, Ohki, M, Wang, Y, Lupala, C.S, Kim, M, Liu, H, Park, S.Y, Lee, W. | | 登録日 | 2019-06-03 | | 公開日 | 2020-06-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The crystal structure of light-driven cyanobacterial chloride importer from Mastigocladopsis repens with Bromide ion
To Be Published
|
|
4H8A
 
 | | Crystal structure of ureidoglycolate dehydrogenase in binary complex with NADH | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Ureidoglycolate dehydrogenase | | 著者 | Rhee, S, Shin, I, Kim, M. | | 登録日 | 2012-09-22 | | 公開日 | 2013-01-16 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Structural and functional insights into (s)-ureidoglycolate dehydrogenase, a metabolic branch point enzyme in nitrogen utilization.
Plos One, 7, 2012
|
|
5CKR
 
 | | Crystal Structure of MraY in complex with Muraymycin D2 | | 分子名称: | Muraymycin D2, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | 著者 | Lee, S.Y, Chung, B.C, Mashalidis, E.H, Tanino, T, Kim, M, Hong, J, Ichikawa, S. | | 登録日 | 2015-07-15 | | 公開日 | 2016-03-02 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Structural insights into inhibition of lipid I production in bacterial cell wall synthesis.
Nature, 533, 2016
|
|
6BSH
 
 | | Structure of HIV-1 RT complexed with RNA/DNA hybrid in the RNA hydrolysis mode | | 分子名称: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, CALCIUM ION, DNA (5'-D(*GP*TP*AP*TP*GP*CP*CP*AP*CP*TP*AP*GP*TP*TP*AP*TP*TP*GP*TP*GP*GP*CP*C)-3'), ... | | 著者 | Tian, L, Kim, M, Yang, W. | | 登録日 | 2017-12-03 | | 公開日 | 2018-01-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.649 Å) | | 主引用文献 | Structure of HIV-1 reverse transcriptase cleaving RNA in an RNA/DNA hybrid.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BSI
 
 | | Structure of HIV-1 RT complexed with an RNA/DNA hybrid containing the polypurine-tract sequence | | 分子名称: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, CALCIUM ION, DNA (5'-D(*GP*TP*TP*TP*TP*TP*CP*TP*TP*TP*TP*GP*TP*TP*AP*TP*TP*GP*TP*GP*GP*CP*C)-3'), ... | | 著者 | Tian, L, Kim, M, Yang, W. | | 登録日 | 2017-12-03 | | 公開日 | 2018-01-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Structure of HIV-1 reverse transcriptase cleaving RNA in an RNA/DNA hybrid.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BSJ
 
 | | Structure of HIV-1 RT complexed with an RNA/DNA hybrid sequence non-preferred for RNA hydrolysis | | 分子名称: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, CALCIUM ION, DNA (5'-D(*GP*TP*AP*TP*GP*CP*CP*TP*AP*TP*AP*GP*TP*TP*AP*TP*TP*GP*TP*GP*GP*CP*C)-3'), ... | | 著者 | Tian, L, Kim, M, Yang, W. | | 登録日 | 2017-12-03 | | 公開日 | 2018-01-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Structure of HIV-1 reverse transcriptase cleaving RNA in an RNA/DNA hybrid.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BSG
 
 | | Structure of HIV-1 RT complexed with RNA/DNA hybrid in an RNA hydrolysis-off mode | | 分子名称: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Tian, L, Kim, M, Yang, W. | | 登録日 | 2017-12-03 | | 公開日 | 2018-01-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | Structure of HIV-1 reverse transcriptase cleaving RNA in an RNA/DNA hybrid.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3CLJ
 
 | | Structure of the RNA polymerase II CTD-interacting domain of Nrd1 | | 分子名称: | GLYCEROL, Protein NRD1, SULFATE ION | | 著者 | Vasiljeva, L, Kim, M, Mutschler, H, Buratowski, S, Meinhart, A. | | 登録日 | 2008-03-19 | | 公開日 | 2008-07-29 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The Nrd1-Nab3-Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain.
Nat.Struct.Mol.Biol., 15, 2008
|
|
3W30
 
 | | Structual basis for the recognition of Ubc13 by the Shigella flexneri effector OspI | | 分子名称: | ORF169b | | 著者 | Nishide, A, Kim, M, Takagi, K, Sasakawa, C, Mizushima, T. | | 登録日 | 2012-12-07 | | 公開日 | 2013-03-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.99 Å) | | 主引用文献 | Structural Basis for the Recognition of Ubc13 by the Shigella flexneri Effector OspI.
J.Mol.Biol., 425, 2013
|
|
3W31
 
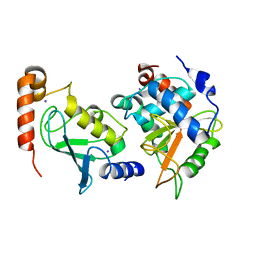 | | Structual basis for the recognition of Ubc13 by the Shigella flexneri effector OspI | | 分子名称: | IODIDE ION, ORF169b, Ubiquitin-conjugating enzyme E2 N | | 著者 | Nishide, A, Kim, M, Takagi, K, Sasakawa, C, Mizushima, T. | | 登録日 | 2012-12-07 | | 公開日 | 2013-03-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.96 Å) | | 主引用文献 | Structural basis for the recognition of Ubc13 by the Shigella flexneri effector OspI.
J.Mol.Biol., 425, 2013
|
|
2KM4
 
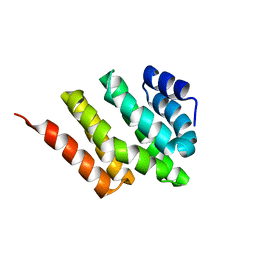 | | Solution structure of Rtt103 CTD interacting domain | | 分子名称: | Regulator of Ty1 transposition protein 103 | | 著者 | Lunde, B.M, Reichow, S, Kim, M, Leeper, T.C, Becker, R, Buratowski, S, Meinhart, A, Varani, G. | | 登録日 | 2009-07-20 | | 公開日 | 2010-09-08 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Cooperative interaction of transcription termination factors with the RNA polymerase II C-terminal domain.
Nat.Struct.Mol.Biol., 17, 2010
|
|
7YA8
 
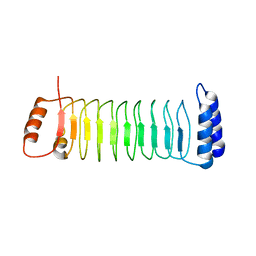 | | The crystal structure of IpaH2.5 LRR domain | | 分子名称: | RING-type E3 ubiquitin transferase | | 著者 | Hiragi, K, Nishide, A, Takagi, K, Iwai, K, Kim, M, Mizushima, T. | | 登録日 | 2022-06-27 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Structural insight into the recognition of the linear ubiquitin assembly complex by Shigella E3 ligase IpaH1.4/2.5.
J.Biochem., 173, 2023
|
|
7YA7
 
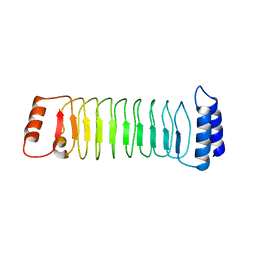 | | The crystal structure of IpaH1.4 LRR domain | | 分子名称: | RING-type E3 ubiquitin transferase | | 著者 | Hiragi, K, Nishide, A, Takagi, K, Iwai, K, Kim, M, Mizushima, T. | | 登録日 | 2022-06-27 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural insight into the recognition of the linear ubiquitin assembly complex by Shigella E3 ligase IpaH1.4/2.5.
J.Biochem., 173, 2023
|
|
