3A5T
 
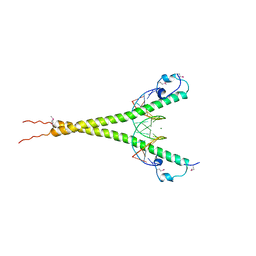 | | Crystal structure of MafG-DNA complex | | Descriptor: | 5'-D(*CP*TP*GP*AP*TP*GP*AP*GP*TP*CP*AP*GP*CP*AP*C)-3', 5'-D(*GP*TP*GP*CP*TP*GP*AP*CP*TP*CP*AP*TP*CP*AP*G)-3', MAGNESIUM ION, ... | | Authors: | Kurokawa, H, Motohashi, H, Sueno, S, Kimura, M, Takagawa, H, Kanno, Y, Yamamoto, M, Tanaka, T. | | Deposit date: | 2009-08-11 | | Release date: | 2009-10-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Alternative DNA Recognition by Maf Transcription Factors
Mol.Cell.Biol., 29, 2009
|
|
3AML
 
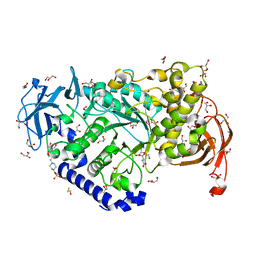 | | Structure of the Starch Branching Enzyme I (BEI) from Oryza sativa L | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | Authors: | Kakuta, Y, Chaen, K, Noguchi, J, Vu, N, Kimura, M. | | Deposit date: | 2010-08-20 | | Release date: | 2011-09-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the branching enzyme I (BEI) from Oryza sativa L with implications for catalysis and substrate binding.
Glycobiology, 21, 2011
|
|
3AMK
 
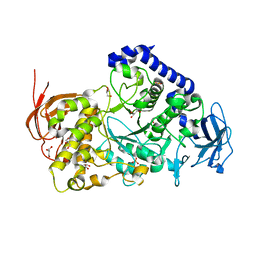 | | Structure of the Starch Branching Enzyme I (BEI) from Oryza sativa L | | Descriptor: | GLYCEROL, Os06g0726400 protein, PHOSPHATE ION | | Authors: | Kakuta, Y, Chaen, K, Noguchi, J, Vu, N, Kimura, M. | | Deposit date: | 2010-08-20 | | Release date: | 2011-09-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the branching enzyme I (BEI) from Oryza sativa L with implications for catalysis and substrate binding.
Glycobiology, 21, 2011
|
|
1J1F
 
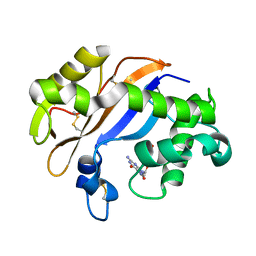 | | Crystal structure of the RNase MC1 mutant N71T in complex with 5'-GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, RIBONUCLEASE MC1 | | Authors: | Numata, T, Suzuki, A, Kakuta, Y, Kimura, K, Yao, M, Tanaka, I, Yoshida, Y, Ueda, T, Kimura, M. | | Deposit date: | 2002-12-03 | | Release date: | 2003-05-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of the Ribonuclease MC1 Mutants N71T and N71S in Complex with 5'-GMP: Structural Basis for Alterations in Substrate Specificity
Biochemistry, 42, 2003
|
|
1J1G
 
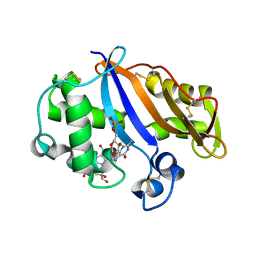 | | Crystal structure of the RNase MC1 mutant N71S in complex with 5'-GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Ribonuclease MC1 | | Authors: | Numata, T, Suzuki, A, Kakuta, Y, Kimura, K, Yao, M, Tanaka, I, Yoshida, Y, Ueda, T, Kimura, M. | | Deposit date: | 2002-12-04 | | Release date: | 2003-05-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of the Ribonuclease MC1 Mutants N71T and N71S in Complex with 5'-GMP: Structural Basis for Alterations in Substrate Specificity
Biochemistry, 42, 2003
|
|
6T1M
 
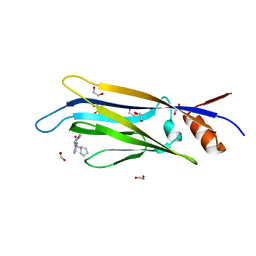 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with benzimidazole-amide derivative 4 | | Descriptor: | 1,2-ETHANEDIOL, 4-cyano-~{N}-[2-(piperidin-1-ylmethyl)-1~{H}-benzimidazol-5-yl]benzamide, Protein ENL | | Authors: | Chaikuad, A, Heidenreich, D, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
6T1N
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with benzimidazole-amide derivative 5 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloranyl-~{N}-[2-(piperidin-1-ylmethyl)-3~{H}-benzimidazol-5-yl]benzamide, Protein ENL | | Authors: | Chaikuad, A, Heidenreich, D, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
6T1O
 
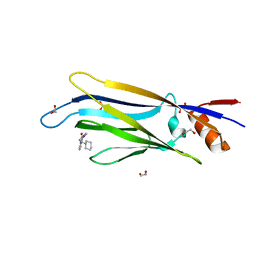 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with benzimidazole-amide derivative 6 | | Descriptor: | 1,2-ETHANEDIOL, 4-iodanyl-~{N}-[2-(piperidin-1-ylmethyl)-3~{H}-benzimidazol-5-yl]benzamide, Protein ENL | | Authors: | Chaikuad, A, Heidenreich, D, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
5NKL
 
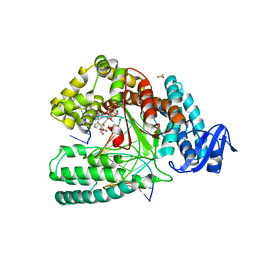 | | Crystal structure of the large fragment of DNA polymerase I from Thermus Aquaticus in a closed ternary complex with the artificial base pair dDs-dPxTP | | Descriptor: | ACETATE ION, DNA (5'-D(*AP*AP*AP*(DNU)P*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), ... | | Authors: | Betz, K, Marx, A, Diederichs, K, Hirao, I, Kimoto, M. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Expansion of the Genetic Alphabet with an Artificial Nucleobase Pair.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
1X0T
 
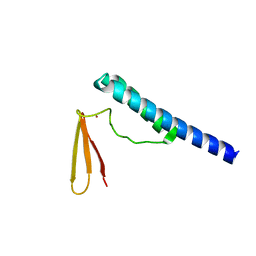 | | Crystal structure of ribonuclease P protein Ph1601p from Pyrococcus horikoshii OT3 | | Descriptor: | Ribonuclease P protein component 4, ZINC ION | | Authors: | Kakuta, Y, Ishimatsu, I, Numata, T, Kimura, K, Yao, M, Tanaka, I, Kimura, M. | | Deposit date: | 2005-03-29 | | Release date: | 2005-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of a Ribonuclease P Protein Ph1601p from Pyrococcus horikoshii OT3: An Archaeal Homologue of Human Nuclear Ribonuclease P Protein Rpp21(,)
Biochemistry, 44, 2005
|
|
1ET7
 
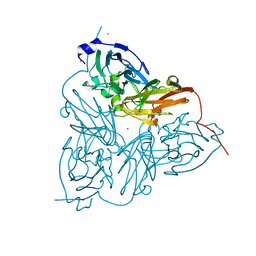 | | CRYSTAL STRUCTURE OF NITRITE REDUCTASE HIS255ASP MUTANT FROM ALCALIGENES FAECALIS S-6 | | Descriptor: | CADMIUM ION, COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Boulanger, M.J, Kukimoto, M, Nishiyama, M, Horinouchi, S, Murphy, M.E.P. | | Deposit date: | 2000-04-12 | | Release date: | 2000-08-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Catalytic roles for two water bridged residues (Asp-98 and His-255) in the active site of copper-containing nitrite reductase.
J.Biol.Chem., 275, 2000
|
|
1ET5
 
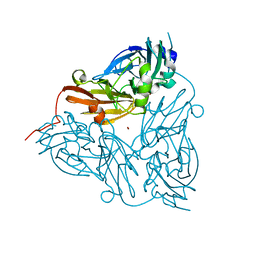 | | CRYSTAL STRUCTURE OF NITRITE REDUCTASE ASP98ASN MUTANT FROM ALCALIGENES FAECALIS S-6 | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE, ZINC ION | | Authors: | Boulanger, M.J, Kukimoto, M, Nishiyama, M, Horinouchi, S, Murphy, M.E.P. | | Deposit date: | 2000-04-12 | | Release date: | 2000-08-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalytic roles for two water bridged residues (Asp-98 and His-255) in the active site of copper-containing nitrite reductase.
J.Biol.Chem., 275, 2000
|
|
1ET8
 
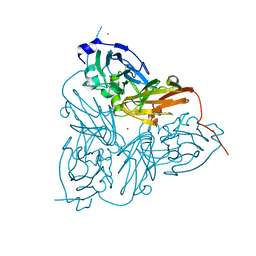 | | CRYSTAL STRUCTURE OF NITRITE REDUCTASE HIS255ASN MUTANT FROM ALCALIGENES FAECALIS | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE, ZINC ION | | Authors: | Boulanger, M.J, Kukimoto, M, Nishiyama, M, Horinouchi, S, Murphy, M.E.P. | | Deposit date: | 2000-04-12 | | Release date: | 2000-08-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Catalytic roles for two water bridged residues (Asp-98 and His-255) in the active site of copper-containing nitrite reductase.
J.Biol.Chem., 275, 2000
|
|
3VSM
 
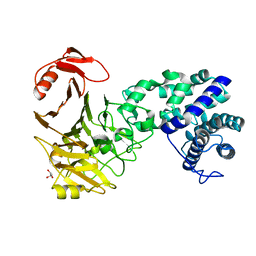 | | The crystal structure of novel chondroition lyase ODV-E66, baculovirus envelope protein | | Descriptor: | GLYCEROL, Occlusion-derived virus envelope protein E66 | | Authors: | Kawaguchi, Y, Sugiura, N, Kimata, K, Kimura, M, Kakuta, Y. | | Deposit date: | 2012-04-27 | | Release date: | 2013-05-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of novel chondroition lyase ODV-E66, baculovirus envelope protein
To be Published
|
|
3VSN
 
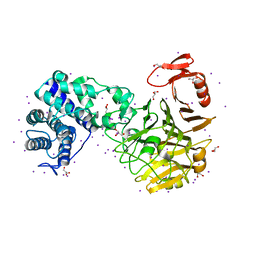 | | The crystal structure of novel chondroition lyase ODV-E66, baculovirus envelope protein | | Descriptor: | GLYCEROL, IODIDE ION, Occlusion-derived virus envelope protein E66 | | Authors: | Kawaguchi, Y, Sugiura, N, Kimata, K, Kimura, M, Kakuta, Y. | | Deposit date: | 2012-04-27 | | Release date: | 2013-05-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of novel chondroition lyase ODV-E66, baculovirus envelope protein
To be Published
|
|
1V9H
 
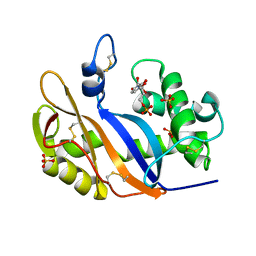 | | Crystal structure of the RNase MC1 mutant Y101A in complex with 5'-UMP | | Descriptor: | Ribonuclease MC, SULFATE ION, URIDINE-5'-MONOPHOSPHATE | | Authors: | Kimura, K, Numata, T, Kakuta, Y, Kimura, M. | | Deposit date: | 2004-01-26 | | Release date: | 2004-10-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Amino acids conserved at the C-terminal half of the ribonuclease t2 family contribute to protein stability of the enzymes
Biosci.Biotechnol.Biochem., 68, 2004
|
|
2Z87
 
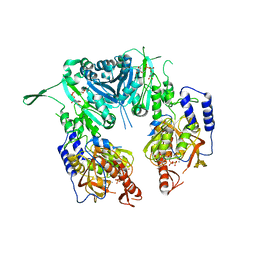 | | Crystal structure of chondroitin polymerase from Escherichia coli strain K4 (K4CP) complexed with UDP-GalNAc and UDP | | Descriptor: | Chondroitin synthase, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Osawa, T, Sugiura, N, Shimada, H, Hirooka, R, Tsuji, A, Kimura, M, Kimata, K, Kakuta, Y. | | Deposit date: | 2007-09-03 | | Release date: | 2008-09-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of chondroitin polymerase from Escherichia coli K4.
Biochem. Biophys. Res. Commun., 378, 2009
|
|
2Z86
 
 | | Crystal structure of chondroitin polymerase from Escherichia coli strain K4 (K4CP) complexed with UDP-GlcUA and UDP | | Descriptor: | Chondroitin synthase, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Osawa, T, Sugiura, N, Shimada, H, Hirooka, R, Tsuji, A, Kimura, M, Kimata, K, Kakuta, Y. | | Deposit date: | 2007-09-03 | | Release date: | 2008-09-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of chondroitin polymerase from Escherichia coli K4.
Biochem. Biophys. Res. Commun., 378, 2009
|
|
7D5R
 
 | | Structure of the Ca2+-bound C646A mutant of peptidylarginine deiminase type III (PAD3) | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Mashimo, R, Akimoto, M, Unno, M. | | Deposit date: | 2020-09-28 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.148 Å) | | Cite: | Structures of human peptidylarginine deiminase type III provide insights into substrate recognition and inhibitor design.
Arch.Biochem.Biophys., 708, 2021
|
|
6HT1
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with SGC-iMLLT (compound 92) | | Descriptor: | 1,2-ETHANEDIOL, 1-methyl-~{N}-[2-[[(2~{S})-2-methylpyrrolidin-1-yl]methyl]-3~{H}-benzimidazol-5-yl]indazole-5-carboxamide, Protein ENL, ... | | Authors: | Heidenreich, D, Chaikuad, A, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-10-02 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of an MLLT1/3 YEATS Domain Chemical Probe.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6HPW
 
 | | Crystal structure of ENL (MLLT1) in complex with compound 20 | | Descriptor: | 1,2-ETHANEDIOL, 3-iodanyl-4-methyl-~{N}-[2-(piperidin-1-ylmethyl)-3~{H}-benzimidazol-5-yl]benzamide, Protein ENL | | Authors: | Heidenreich, D, Chaikuad, A, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Approach toward Identification of Inhibitory Fragments for Eleven-Nineteen-Leukemia Protein (ENL).
J.Med.Chem., 61, 2018
|
|
6HT0
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with compound 94 | | Descriptor: | 1,2-ETHANEDIOL, 1-cyclopropyl-~{N}-[2-[[(2~{S})-2-methylpyrrolidin-1-yl]methyl]-3~{H}-benzimidazol-5-yl]indazole-5-carboxamide, Protein ENL, ... | | Authors: | Heidenreich, D, Chaikuad, A, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-10-02 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of an MLLT1/3 YEATS Domain Chemical Probe.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
1WN5
 
 | | Crystal Structure of Blasticidin S Deaminase (BSD) Complexed with Cacodylic Acid | | Descriptor: | Blasticidin-S deaminase, CACODYLATE ION, ZINC ION | | Authors: | Kumasaka, T, Yamamoto, M, Furuichi, M, Nakasako, M, Kimura, M, Yamaguchi, I, Ueki, T. | | Deposit date: | 2004-07-27 | | Release date: | 2005-10-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of blasticidin S deaminase (BSD): implications for dynamic properties of catalytic zinc
J.Biol.Chem., 282, 2007
|
|
7CG3
 
 | | Staggered ring conformation of CtHsp104 (Hsp104 from Chaetomium Thermophilum) | | Descriptor: | Heat shock protein 104 | | Authors: | Inoue, Y, Hanazono, Y, Noi, K, Kawamoto, A, Kimatsuka, M, Harada, R, Takeda, K, Iwamasa, N, Shibata, K, Noguchi, K, Shigeta, Y, Namba, K, Ogura, T, Miki, K, Shinohara, K, Yohda, M. | | Deposit date: | 2020-06-30 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Split conformation of Chaetomium thermophilum Hsp104 disaggregase.
Structure, 29, 2021
|
|
1WN6
 
 | | Crystal Structure of Blasticidin S Deaminase (BSD) Complexed with Tetrahedral Intermediate of Blasticidin S | | Descriptor: | 6-(4-AMINO-4-HYDROXY-2-OXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-3-[3-AMINO-5-(N-METHYL-GUANIDINO)-PENT ANOYLAMINO]-3,6-DIHYDRO-2H-PYRAN-2-CARBOXYLIC ACID, ARSENIC, Blasticidin-S deaminase, ... | | Authors: | Kumasaka, T, Yamamoto, M, Furuichi, M, Nakasako, M, Kimura, M, Yamaguchi, I, Ueki, T. | | Deposit date: | 2004-07-27 | | Release date: | 2005-10-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of blasticidin S deaminase (BSD): implications for dynamic properties of catalytic zinc
J.Biol.Chem., 282, 2007
|
|
