4TN5
 
 | |
2B7Q
 
 | | Crystal structure of quinolinic acid phosphoribosyltransferase from Helicobacter pylori with nicotinate mononucleotide | | Descriptor: | NICOTINATE MONONUCLEOTIDE, Probable nicotinate-nucleotide pyrophosphorylase | | Authors: | Kim, M.K, Im, Y.J, Lee, J.H, Eom, S.H. | | Deposit date: | 2005-10-05 | | Release date: | 2006-02-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Crystal structure of quinolinic acid phosphoribosyltransferase from Helicobacter pylori
Proteins, 63, 2006
|
|
2B7P
 
 | | Crystal structure of quinolinic acid phosphoribosyltransferase from Helicobacter pylori with phthalic acid | | Descriptor: | PHTHALIC ACID, Probable nicotinate-nucleotide pyrophosphorylase, SULFATE ION | | Authors: | Kim, M.K, Im, Y.J, Lee, J.H, Eom, S.H. | | Deposit date: | 2005-10-05 | | Release date: | 2006-02-14 | | Last modified: | 2018-09-19 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of quinolinic acid phosphoribosyltransferase from Helicobacter pylori
Proteins, 63, 2006
|
|
2B7N
 
 | | Crystal structure of quinolinic acid phosphoribosyltransferase from Helicobacter pylori | | Descriptor: | Probable nicotinate-nucleotide pyrophosphorylase, QUINOLINIC ACID, SULFATE ION | | Authors: | Kim, M.K, Im, Y.J, Lee, J.H, Eom, S.H. | | Deposit date: | 2005-10-04 | | Release date: | 2006-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of quinolinic acid phosphoribosyltransferase from Helicobacter pylori
Proteins, 63, 2006
|
|
1IVW
 
 | | Crystal structure of copper amine oxidase from Arthrobacter globiformis: Late intermediate in topaquinone biogenesis | | Descriptor: | COPPER (II) ION, amine oxidase | | Authors: | Kim, M, Okajima, T, Kishishita, S, Yoshimura, M, Kawamori, A, Tanizawa, K, Yamaguchi, H. | | Deposit date: | 2002-03-29 | | Release date: | 2002-08-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray snapshots of quinone cofactor biogenesis in bacterial copper amine oxidase.
Nat.Struct.Biol., 9, 2002
|
|
1IVX
 
 | | Crystal structure of copper amine oxidase from Arthrobacter globiformis: Holo form generated by biogenesis in crystal. | | Descriptor: | COPPER (II) ION, amine oxidase | | Authors: | Kim, M, Okajima, T, Kishishita, S, Yoshimura, M, Kawamori, A, Tanizawa, K, Yamaguchi, H. | | Deposit date: | 2002-03-29 | | Release date: | 2002-08-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray snapshots of quinone cofactor biogenesis in bacterial copper amine oxidase.
Nat.Struct.Biol., 9, 2002
|
|
1IVU
 
 | | Crystal structure of copper amine oxidase from Arthrobacter globiformis: Initial intermediate in topaquinone biogenesis | | Descriptor: | COPPER (II) ION, amine oxidase | | Authors: | Kim, M, Okajima, T, Kishishita, S, Yoshimura, M, Kawamori, A, Tanizawa, K, Yamaguchi, H. | | Deposit date: | 2002-03-29 | | Release date: | 2002-08-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray snapshots of quinone cofactor biogenesis in bacterial copper amine oxidase.
Nat.Struct.Biol., 9, 2002
|
|
1IVV
 
 | | Crystal structure of copper amine oxidase from Arthrobacter globiformis: Early intermediate in topaquinone biogenesis | | Descriptor: | COPPER (II) ION, amine oxidase | | Authors: | Kim, M, Okajima, T, Kishishita, S, Yoshimura, M, Kawamori, A, Tanizawa, K, Yamaguchi, H. | | Deposit date: | 2002-03-29 | | Release date: | 2002-08-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray snapshots of quinone cofactor biogenesis in bacterial copper amine oxidase.
Nat.Struct.Biol., 9, 2002
|
|
4J4K
 
 | | Crystal structure of glucose isomerase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Xylose isomerase, ... | | Authors: | Kim, M.K, An, Y.J, Lee, S, Jeong, C.S, Cha, S.S. | | Deposit date: | 2013-02-07 | | Release date: | 2014-04-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of glucose isomerase
To be Published
|
|
4GQX
 
 | | Crystal structure of EIIA(NTR) from Burkholderia pseudomallei | | Descriptor: | PTS IIA-like nitrogen-regulatory protein PtsN | | Authors: | Kim, M.-S, Shin, D.H. | | Deposit date: | 2012-08-24 | | Release date: | 2013-03-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | New molecular interaction of IIA(Ntr) and HPr from Burkholderia pseudomallei identified by X-ray crystallography and docking studies
Proteins, 81, 2013
|
|
4MB2
 
 | | Crystal structure of TON1374 in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Phosphopantothenate synthetase | | Authors: | Kim, M.-K, An, Y.J, Cha, S.-S. | | Deposit date: | 2013-08-19 | | Release date: | 2014-08-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The crystal structure of a novel phosphopantothenate synthetase from the hyperthermophilic archaea, Thermococcus onnurineus NA1
Biochem.Biophys.Res.Commun., 439, 2013
|
|
6JFV
 
 | | The crystal structure of 2B-2B complex from keratins 5 and 14 (C367A mutant of K14) | | Descriptor: | Keratin, type I cytoskeletal 14, type II cytoskeletal 5 | | Authors: | Kim, M.S, Lee, C.H, Coulombe, P.A, Leahy, D.J. | | Deposit date: | 2019-02-12 | | Release date: | 2020-01-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Function Analyses of a Keratin Heterotypic Complex Identify Specific Keratin Regions Involved in Intermediate Filament Assembly.
Structure, 28, 2020
|
|
7XC0
 
 | | Crystal structure of Human RPTPH | | Descriptor: | PHOSPHATE ION, Receptor-type tyrosine-protein phosphatase H | | Authors: | Kim, M, Ryu, S.E. | | Deposit date: | 2022-03-22 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structure of the catalytic domain of human RPTPH.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
1MG4
 
 | | STRUCTURE OF N-TERMINAL DOUBLECORTIN DOMAIN FROM DCLK: WILD TYPE PROTEIN | | Descriptor: | DOUBLECORTIN-LIKE KINASE (N-TERMINAL DOMAIN), SULFATE ION | | Authors: | Kim, M.H, Cierpickil, T, Derewenda, U, Krowarsch, D, Feng, Y, Devedjiev, Y, Dauter, Z, Walsh, C.A, Otlewski, J, Bushweller, J.H, Derewenda, Z. | | Deposit date: | 2002-08-14 | | Release date: | 2003-04-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.504 Å) | | Cite: | The DCX-domain Tandems of Doublecortin and Doublecortin-like Kinase
Nat.Struct.Biol., 10, 2003
|
|
1MFW
 
 | | STRUCTURE OF N-TERMINAL DOUBLECORTIN DOMAIN FROM DCLK: SELENOMETHIONINE LABELED PROTEIN | | Descriptor: | DOUBLECORTIN-LIKE KINASE (N-TERMINAL DOMAIN), SULFATE ION | | Authors: | Kim, M.H, Cierpickil, T, Derewenda, U, Krowarsch, D, Feng, Y, Devedjiev, Y, Dauter, Z, Walsh, C.A, Otlewski, J, Bushweller, J.H, Derewenda, Z. | | Deposit date: | 2002-08-13 | | Release date: | 2003-04-29 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The DCX-domain Tandems of Doublecortin and Doublecortin-like Kinase
Nat.Struct.Biol., 10, 2003
|
|
4MB0
 
 | | Crystal structure of TON1374 | | Descriptor: | ACETATE ION, phosphopantothenate synthetase | | Authors: | Kim, M.-K, An, Y.J, Cha, S.-S. | | Deposit date: | 2013-08-19 | | Release date: | 2014-08-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The crystal structure of a novel phosphopantothenate synthetase from the hyperthermophilic archaea, Thermococcus onnurineus NA1
Biochem.Biophys.Res.Commun., 439, 2013
|
|
5ZQH
 
 | | Crystal structure of Streptococcus transcriptional regulator | | Descriptor: | PadR family transcriptional regulator | | Authors: | Kim, M, Hong, M. | | Deposit date: | 2018-04-19 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based functional analysis of a PadR transcription factor from Streptococcus pneumoniae and characteristic features in the PadR subfamily-2.
Biochem.Biophys.Res.Commun., 532, 2020
|
|
6A7H
 
 | | Bacterial protein toxins | | Descriptor: | RTX toxin, SULFATE ION | | Authors: | Kim, M.H, Hwang, J, Jang, S.Y. | | Deposit date: | 2018-07-03 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural basis of inactivation of Ras and Rap1 small GTPases by Ras/Rap1-specific endopeptidase from the sepsis-causing pathogenVibrio vulnificus
J. Biol. Chem., 293, 2018
|
|
5ZE2
 
 | | Hairpin Complex, RAG1/2-hairpin 12RSS/23RSS complex in 5mM Mn2+ for 2 min at 4'C | | Descriptor: | 1,2-ETHANEDIOL, DNA (30-MER), DNA (31-MER), ... | | Authors: | Kim, M.S, Chuenchor, W, Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2018-02-25 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination
Mol. Cell, 70, 2018
|
|
5ZE1
 
 | | Hairpin Forming Complex, RAG1/2-Nicked 12RSS/23RSS complex in 2mM Mn2+ for 10 min at 4'C | | Descriptor: | 1,2-ETHANEDIOL, DNA, HMGB1 A-B box, ... | | Authors: | Kim, M.S, Chuenchor, W, Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2018-02-25 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination
Mol. Cell, 70, 2018
|
|
5ZDZ
 
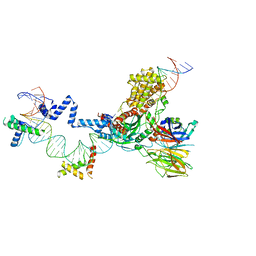 | | Hairpin Forming Complex, RAG1/2-Nicked 12RSS/23RSS complex in Ca2+ | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DNA (30-MER), ... | | Authors: | Kim, M.S, Chuenchor, W, Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2018-02-25 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination
Mol. Cell, 70, 2018
|
|
5ZE0
 
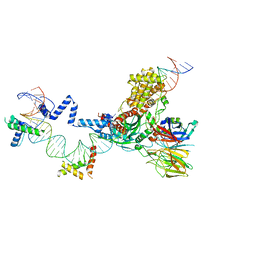 | | Hairpin Forming Complex, RAG1/2-Nicked(with Dideoxy) 12RSS/23RSS complex in Mg2+ | | Descriptor: | 1,2-ETHANEDIOL, DNA (30-MER), DNA (39-MER), ... | | Authors: | Kim, M.S, Chuenchor, W, Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2018-02-25 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination
Mol. Cell, 70, 2018
|
|
5Z7Q
 
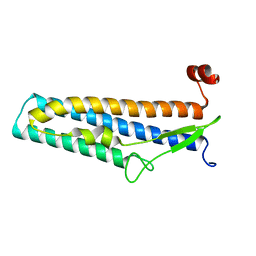 | | Crystal structure of Bacillus cereus flagellin | | Descriptor: | Flagellin | | Authors: | Kim, M, Hong, M. | | Deposit date: | 2018-01-30 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Bacillus cereus flagellin and structure-guided fusion-protein designs
Sci Rep, 8, 2018
|
|
5ZXN
 
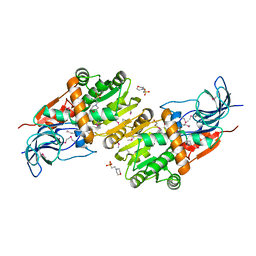 | | Crystal structure of CurA from Vibrio vulnificus | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, NADP-dependent oxidoreductase | | Authors: | Kim, M.-K, Bae, D.-W, Cha, S.-S. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (1.855 Å) | | Cite: | Structural and Biochemical Characterization of the Curcumin-Reducing Activity of CurA from Vibrio vulnificus.
J. Agric. Food Chem., 66, 2018
|
|
5ZXU
 
 | |
