6IZE
 
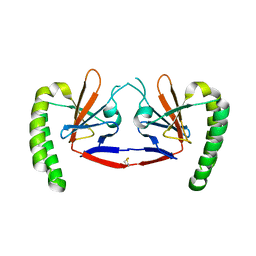 | | Dimeric human TCTP | | Descriptor: | Translationally-controlled tumor protein | | Authors: | Shin, D.H, Kim, M.S. | | Deposit date: | 2018-12-19 | | Release date: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Dimeric human TCTP
To Be Published
|
|
3TT7
 
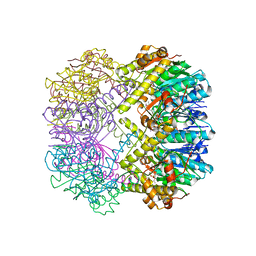 | |
3TT6
 
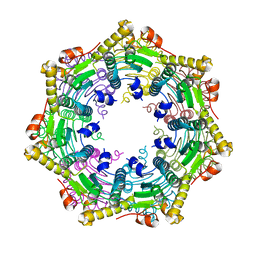 | |
3MCD
 
 | | Crystal structure of Helicobacter pylori MinE, a cell division topological specificity factor | | Descriptor: | Cell division topological specificity factor | | Authors: | Kang, G.B, Song, H.E, Kim, M.K, Youn, H.S, Lee, J.G, An, J.Y, Jeon, H, Chun, J.S, Eom, S.H. | | Deposit date: | 2010-03-29 | | Release date: | 2010-05-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of Helicobacter pylori MinE, a cell division topological specificity factor
Mol.Microbiol., 76, 2010
|
|
2PV6
 
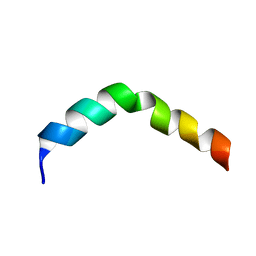 | | HIV-1 gp41 Membrane Proximal Ectodomain Region peptide in DPC micelle | | Descriptor: | Envelope glycoprotein | | Authors: | Sun, Z.-Y.J, Oh, K.J, Kim, M, Reinherz, E.L, Wagner, G. | | Deposit date: | 2007-05-09 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | HIV-1 broadly neutralizing antibody extracts its epitope from a kinked gp41 ectodomain region on the viral membrane
Immunity, 28, 2008
|
|
6IZB
 
 | |
5XLN
 
 | |
5Y7D
 
 | | Crystal structure of human Endothelial-overexpressed LPS associated factor 1 | | Descriptor: | CHLORIDE ION, GLYCEROL, Protein CXorf40A, ... | | Authors: | Park, S.H, Kim, M.J, Park, J.S, Kim, H.J, Han, B.W. | | Deposit date: | 2017-08-17 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal Structure of Human EOLA1 Implies Its Possibility of RNA Binding.
Molecules, 24, 2019
|
|
5Y16
 
 | | Crystal structure of human DUSP28(Y102H) | | Descriptor: | CHLORIDE ION, Dual specificity phosphatase 28, PHOSPHATE ION | | Authors: | Ku, B, Kim, M, Kim, S.J, Ryu, S.E. | | Deposit date: | 2017-07-19 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structural and biochemical analysis of atypically low dephosphorylating activity of human dual-specificity phosphatase 28
PLoS ONE, 12, 2017
|
|
5X3R
 
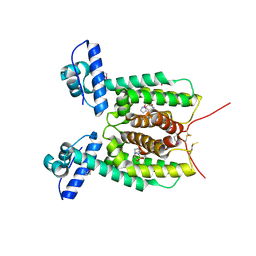 | | Crystal structure of the SmcR complexed with QStatin | | Descriptor: | 1-(5-bromanylthiophen-2-yl)sulfonylpyrazole, LuxR family transcriptional regulator, SULFATE ION | | Authors: | Jang, S.Y, Hwang, J, Kim, M.H. | | Deposit date: | 2017-02-07 | | Release date: | 2018-01-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | QStatin, a Selective Inhibitor of Quorum Sensing inVibrioSpecies
MBio, 9, 2018
|
|
5Z0B
 
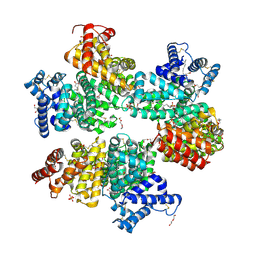 | |
3VSM
 
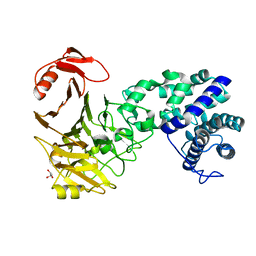 | | The crystal structure of novel chondroition lyase ODV-E66, baculovirus envelope protein | | Descriptor: | GLYCEROL, Occlusion-derived virus envelope protein E66 | | Authors: | Kawaguchi, Y, Sugiura, N, Kimata, K, Kimura, M, Kakuta, Y. | | Deposit date: | 2012-04-27 | | Release date: | 2013-05-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of novel chondroition lyase ODV-E66, baculovirus envelope protein
To be Published
|
|
1ET5
 
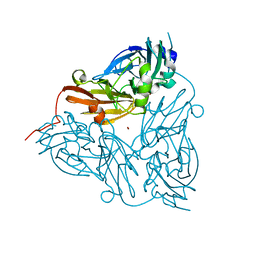 | | CRYSTAL STRUCTURE OF NITRITE REDUCTASE ASP98ASN MUTANT FROM ALCALIGENES FAECALIS S-6 | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE, ZINC ION | | Authors: | Boulanger, M.J, Kukimoto, M, Nishiyama, M, Horinouchi, S, Murphy, M.E.P. | | Deposit date: | 2000-04-12 | | Release date: | 2000-08-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalytic roles for two water bridged residues (Asp-98 and His-255) in the active site of copper-containing nitrite reductase.
J.Biol.Chem., 275, 2000
|
|
1ET8
 
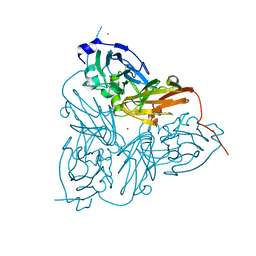 | | CRYSTAL STRUCTURE OF NITRITE REDUCTASE HIS255ASN MUTANT FROM ALCALIGENES FAECALIS | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE, ZINC ION | | Authors: | Boulanger, M.J, Kukimoto, M, Nishiyama, M, Horinouchi, S, Murphy, M.E.P. | | Deposit date: | 2000-04-12 | | Release date: | 2000-08-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Catalytic roles for two water bridged residues (Asp-98 and His-255) in the active site of copper-containing nitrite reductase.
J.Biol.Chem., 275, 2000
|
|
1EJ2
 
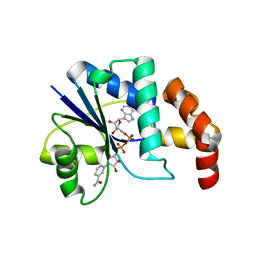 | | Crystal structure of methanobacterium thermoautotrophicum nicotinamide mononucleotide adenylyltransferase with bound NAD+ | | Descriptor: | NICOTINAMIDE MONONUCLEOTIDE ADENYLYLTRANSFERASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Saridakis, V, Christendat, D, Kimber, M.S, Edwards, A.M, Pai, E.F, Midwest Center for Structural Genomics (MCSG), Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2000-02-29 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into ligand binding and catalysis of a central step in NAD+ synthesis: structures of Methanobacterium thermoautotrophicum NMN adenylyltransferase complexes.
J.Biol.Chem., 276, 2001
|
|
3VK9
 
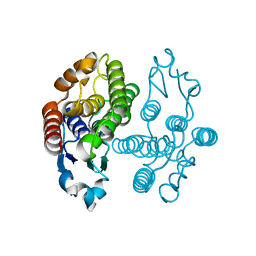 | | Crystal structure of delta-class glutathione transferase from silkmoth | | Descriptor: | GLYCEROL, Glutathione S-transferase delta | | Authors: | Kakuta, Y, Usuda, K, Higashiura, A, Suzuki, M, Nakagawa, A, Kimura, M, Yamamoto, K. | | Deposit date: | 2011-11-10 | | Release date: | 2012-10-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural basis for catalytic activity of a silkworm Delta-class glutathione transferase
Biochim.Biophys.Acta, 1820, 2012
|
|
3VVU
 
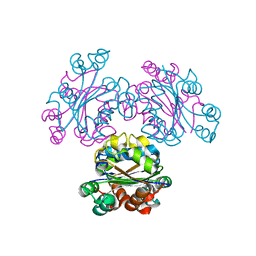 | | Crystal structure of reconstructed bacterial ancestral NDK, Bac1 | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Nemoto, N, Miyazono, K, Kimura, M, Yokobori, S, Akanuma, S, Tanokura, M, Yamagishi, A. | | Deposit date: | 2012-07-27 | | Release date: | 2013-06-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Experimental evidence for the thermophilicity of ancestral life
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3VVT
 
 | | Crystal structure of reconstructed archaeal ancestral NDK, Arc1 | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Nemoto, N, Miyazono, K, Kimura, M, Yokobori, S, Akanuma, S, Tanokura, M, Yamagishi, A. | | Deposit date: | 2012-07-27 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Experimental evidence for the thermophilicity of ancestral life
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4FXQ
 
 | | Full-length Certhrax toxin from Bacillus cereus in complex with Inhibitor P6 | | Descriptor: | 8-fluoro-2-(3-piperidin-1-ylpropanoyl)-1,3,4,5-tetrahydrobenzo[c][1,6]naphthyridin-6(2H)-one, CHLORIDE ION, Putative ADP-ribosyltransferase Certhrax, ... | | Authors: | Visschedyk, D.D, Dimov, S, Kimber, M.S, Park, H.W, Merrill, A.R. | | Deposit date: | 2012-07-03 | | Release date: | 2012-09-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9599 Å) | | Cite: | Certhrax Toxin, an Anthrax-related ADP-ribosyltransferase from Bacillus cereus.
J.Biol.Chem., 287, 2012
|
|
1VCZ
 
 | |
5HNP
 
 | | The structure of the kdo-capped saccharide binding subunit of the O-12 specific ABC transporter, Wzt | | Descriptor: | ABC transporter, CHLORIDE ION | | Authors: | Mallette, E, Mann, E, Whitfield, C, Kimber, M.S. | | Deposit date: | 2016-01-18 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Klebsiella pneumoniae O12 ATP-binding Cassette (ABC) Transporter Recognizes the Terminal Residue of Its O-antigen Polysaccharide Substrate.
J.Biol.Chem., 291, 2016
|
|
7D5R
 
 | | Structure of the Ca2+-bound C646A mutant of peptidylarginine deiminase type III (PAD3) | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Mashimo, R, Akimoto, M, Unno, M. | | Deposit date: | 2020-09-28 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.148 Å) | | Cite: | Structures of human peptidylarginine deiminase type III provide insights into substrate recognition and inhibitor design.
Arch.Biochem.Biophys., 708, 2021
|
|
5L39
 
 | |
8CSF
 
 | | WbbB D232C-Kdo adduct + alpha-Rha(1,3)GlcNAc ternary complex | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, CYTIDINE-5'-MONOPHOSPHATE, N-acetyl glucosaminyl transferase, ... | | Authors: | Forrester, T.J.B, Kimber, M.S. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The retaining beta-Kdo glycosyltransferase WbbB uses a double-displacement mechanism with an intermediate adduct rearrangement step.
Nat Commun, 13, 2022
|
|
8CSE
 
 | | WbbB in complex with alpha-Rha-(1-3)-beta-GlcNAc acceptor | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, N-(8-hydroxyoctyl)-4-methoxybenzamide, N-acetyl glucosaminyl transferase, ... | | Authors: | Forrester, T.J.B, Kimber, M.S. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The retaining beta-Kdo glycosyltransferase WbbB uses a double-displacement mechanism with an intermediate adduct rearrangement step.
Nat Commun, 13, 2022
|
|
