4X3N
 
 | | Crystal structure of 34 kDa F-actin bundling protein from Dictyostelium discoideum | | Descriptor: | CALCIUM ION, CITRIC ACID, Calcium-regulated actin-bundling protein | | Authors: | Kim, M.-K, Kim, J.-H, Kim, J.-S, Kang, S.-O. | | Deposit date: | 2014-12-01 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure of the 34 kDa F-actin-bundling protein ABP34 from Dictyostelium discoideum.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8JCT
 
 | |
2G97
 
 | |
3CZL
 
 | |
3CZK
 
 | | Crystal Structure Analysis of Sucrose hydrolase(SUH) E322Q-sucrose complex | | Descriptor: | Sucrose hydrolase, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Kim, M.I, Rhee, S. | | Deposit date: | 2008-04-29 | | Release date: | 2008-07-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures and mutagenesis of sucrose hydrolase from Xanthomonas axonopodis pv. glycines: insight into the exclusively hydrolytic amylosucrase fold.
J.Mol.Biol., 380, 2008
|
|
1IVU
 
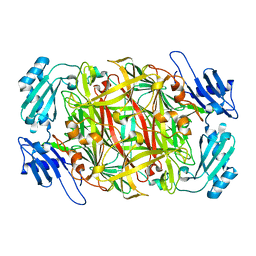 | | Crystal structure of copper amine oxidase from Arthrobacter globiformis: Initial intermediate in topaquinone biogenesis | | Descriptor: | COPPER (II) ION, amine oxidase | | Authors: | Kim, M, Okajima, T, Kishishita, S, Yoshimura, M, Kawamori, A, Tanizawa, K, Yamaguchi, H. | | Deposit date: | 2002-03-29 | | Release date: | 2002-08-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray snapshots of quinone cofactor biogenesis in bacterial copper amine oxidase.
Nat.Struct.Biol., 9, 2002
|
|
1IVV
 
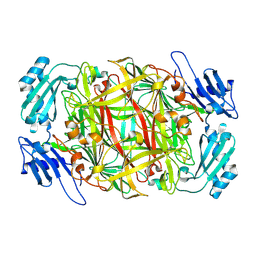 | | Crystal structure of copper amine oxidase from Arthrobacter globiformis: Early intermediate in topaquinone biogenesis | | Descriptor: | COPPER (II) ION, amine oxidase | | Authors: | Kim, M, Okajima, T, Kishishita, S, Yoshimura, M, Kawamori, A, Tanizawa, K, Yamaguchi, H. | | Deposit date: | 2002-03-29 | | Release date: | 2002-08-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray snapshots of quinone cofactor biogenesis in bacterial copper amine oxidase.
Nat.Struct.Biol., 9, 2002
|
|
4GQX
 
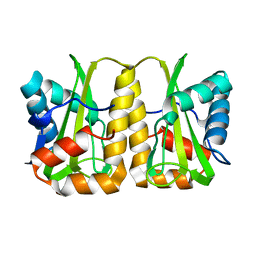 | | Crystal structure of EIIA(NTR) from Burkholderia pseudomallei | | Descriptor: | PTS IIA-like nitrogen-regulatory protein PtsN | | Authors: | Kim, M.-S, Shin, D.H. | | Deposit date: | 2012-08-24 | | Release date: | 2013-03-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | New molecular interaction of IIA(Ntr) and HPr from Burkholderia pseudomallei identified by X-ray crystallography and docking studies
Proteins, 81, 2013
|
|
7X7H
 
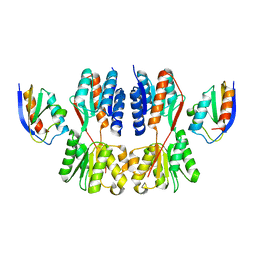 | | Crystal structure of Fructose regulator/Histidine phosphocarrier protein complex from Vibrio cholerae | | Descriptor: | CALCIUM ION, Catabolite repressor/activator, HPr family phosphocarrier protein | | Authors: | Kim, M.-K, Zhang, J, Yoon, C.-K, Seok, Y.-J. | | Deposit date: | 2022-03-09 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | HPr prevents FruR-mediated facilitation of RNA polymerase binding to the fru promoter in Vibrio cholerae.
Nucleic Acids Res., 51, 2023
|
|
2UZ0
 
 | | The Crystal crystal structure of the estA protein, a virulence factor estA protein from Streptococcus pneumonia | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Kim, M.H, Kang, B.S, Kim, K.J, Oh, T.K. | | Deposit date: | 2007-04-23 | | Release date: | 2007-10-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of the Esta Protein, a Virulence Factor from Streptococcus Pneumoniae.
Proteins, 70, 2008
|
|
2C0J
 
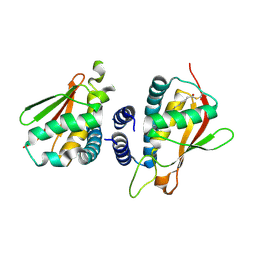 | | Crystal structure of the bet3-trs33 heterodimer | | Descriptor: | PALMITIC ACID, R32611_2, TRAFFICKING PROTEIN PARTICLE COMPLEX SUBUNIT 3 | | Authors: | Kim, M.-S, Yi, M.-J, Lee, K.-H, Wagner, J, Munger, C, Kim, Y.-G, Whiteway, M, Cygler, M, Oh, B.-H, Sacher, M. | | Deposit date: | 2005-09-03 | | Release date: | 2006-02-07 | | Last modified: | 2015-01-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical and Crystallographic Studies Reveal a Specific Interaction between Trapp Subunits Trs33P and Bet3P
Traffic, 6, 2005
|
|
2BR6
 
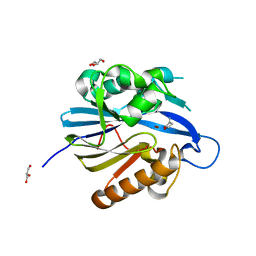 | | Crystal Structure of Quorum-Quenching N-Acyl Homoserine Lactone Lactonase | | Descriptor: | AIIA-LIKE PROTEIN, GLYCEROL, HOMOSERINE LACTONE, ... | | Authors: | Kim, M.H, Choi, W.C, Kang, H.O, Kang, B.S, Kim, K.J, Derewenda, Z.S, Lee, J.K, Oh, T.K, Lee, C.H. | | Deposit date: | 2005-05-03 | | Release date: | 2005-12-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Molecular Structure and Catalytic Mechanism of a Quorum-Quenching N-Acyl-L-Homoserine Lactone Hydrolase.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BTN
 
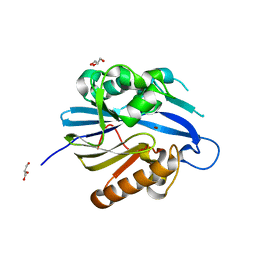 | | Crystal Structure and Catalytic Mechanism of the Quorum-Quenching N- Acyl Homoserine Lactone Hydrolase | | Descriptor: | AIIA-LIKE PROTEIN, GLYCEROL, ZINC ION | | Authors: | Kim, M.H, Choi, W.C, Kang, H.O, Lee, J.S, Kang, B.S, Kim, K.J, Derewenda, Z.S, Oh, T.K, Lee, C.H, Lee, J.K. | | Deposit date: | 2005-06-03 | | Release date: | 2005-12-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Molecular Structure and Catalytic Mechanism of a Quorum-Quenching N-Acyl-L-Homoserine Lactone Hydrolase.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
6IMP
 
 | |
3KZ9
 
 | | Crystal structure of the master transcriptional regulator, SmcR, in Vibrio vulnificus provides insight into its DNA recognition mechanism | | Descriptor: | SULFATE ION, SmcR | | Authors: | Kim, M.H, Kim, Y, Choi, W.-C, Hwang, J. | | Deposit date: | 2009-12-08 | | Release date: | 2010-03-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of SmcR, a quorum-sensing master regulator of Vibrio vulnificus, provides insight into its regulation of transcription
J.Biol.Chem., 285, 2010
|
|
5ZQH
 
 | | Crystal structure of Streptococcus transcriptional regulator | | Descriptor: | PadR family transcriptional regulator | | Authors: | Kim, M, Hong, M. | | Deposit date: | 2018-04-19 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based functional analysis of a PadR transcription factor from Streptococcus pneumoniae and characteristic features in the PadR subfamily-2.
Biochem.Biophys.Res.Commun., 532, 2020
|
|
1GVI
 
 | | Thermus maltogenic amylase in complex with beta-CD | | Descriptor: | Cycloheptakis-(1-4)-(alpha-D-glucopyranose), MALTOGENIC AMYLASE | | Authors: | Kim, M.-S, Kim, J.-I, Oh, B.-H. | | Deposit date: | 2002-02-14 | | Release date: | 2002-06-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Cyclomaltodextrinase, Neopullulanase, and Maltogenic Amylase are Nearly Indistinguishable from Each Other
J.Biol.Chem., 277, 2002
|
|
5IE9
 
 | |
2G95
 
 | |
2GQT
 
 | |
3IO4
 
 | | Huntingtin amino-terminal region with 17 Gln residues - Crystal C90 | | Descriptor: | CALCIUM ION, Maltose-binding periplasmic protein,Huntingtin fusion protein, ZINC ION | | Authors: | Kim, M.W, Chelliah, Y, Kim, S.W, Otwinowski, Z, Bezprozvanny, I. | | Deposit date: | 2009-08-13 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.63 Å) | | Cite: | Secondary structure of Huntingtin amino-terminal region.
Structure, 17, 2009
|
|
3IO6
 
 | | Huntingtin amino-terminal region with 17 Gln residues - crystal C92-a | | Descriptor: | CALCIUM ION, Maltose-binding periplasmic protein, HUNTINGTIN FUSION PROTEIN, ... | | Authors: | Kim, M.W, Chelliah, Y, Kim, S.W, Otwinowski, Z, Bezprozvanny, I. | | Deposit date: | 2009-08-13 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Secondary structure of Huntingtin amino-terminal region.
Structure, 17, 2009
|
|
6A7H
 
 | | Bacterial protein toxins | | Descriptor: | RTX toxin, SULFATE ION | | Authors: | Kim, M.H, Hwang, J, Jang, S.Y. | | Deposit date: | 2018-07-03 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural basis of inactivation of Ras and Rap1 small GTPases by Ras/Rap1-specific endopeptidase from the sepsis-causing pathogenVibrio vulnificus
J. Biol. Chem., 293, 2018
|
|
6A8J
 
 | | Crystal structure of bacterial protein toxins | | Descriptor: | GLYCEROL, RTX toxin, SULFATE ION | | Authors: | Kim, M.H, Hwang, J, Jang, S.Y. | | Deposit date: | 2018-07-09 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.711 Å) | | Cite: | Structural basis of inactivation of Ras and Rap1 small GTPases by Ras/Rap1-specific endopeptidase from the sepsis-causing pathogenVibrio vulnificus
J. Biol. Chem., 293, 2018
|
|
7WG4
 
 | | DVAA-KlAte1 | | Descriptor: | Arginyltransferase, ZINC ION | | Authors: | Kim, M.K, Kim, B.H, Oh, S.-J, Song, H.K. | | Deposit date: | 2021-12-28 | | Release date: | 2022-09-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structure of the Ate1 arginyl-tRNA-protein transferase and arginylation of N-degron substrates.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
