5Y6U
 
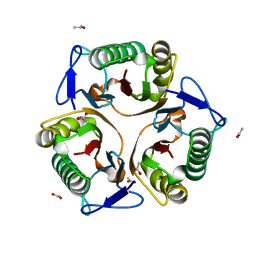 | |
7CD2
 
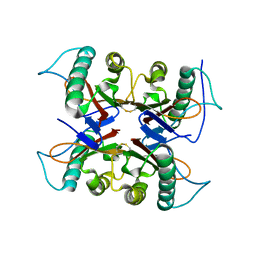 | |
7CD3
 
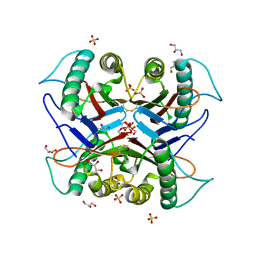 | |
7CD4
 
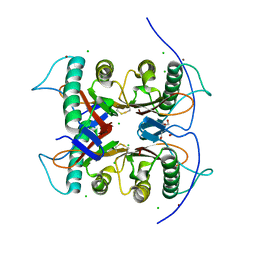 | | Crystal structure of the S103F mutant of Bacillus subtilis (natto) YabJ protein. | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Fujimoto, Z, Kishine, N, Kimura, K. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Tetramer formation of Bacillus subtilis YabJ protein that belongs to YjgF/YER057c/UK114 family.
Biosci.Biotechnol.Biochem., 85, 2021
|
|
5ZFS
 
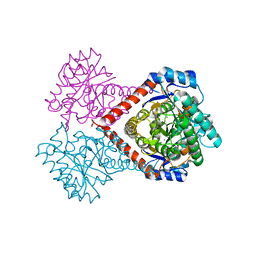 | | Crystal structure of Arthrobacter globiformis M30 sugar epimerase which can produce D-allulose from D-fructose | | Descriptor: | ACETATE ION, D-allulose-3-epimerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Yoshihara, A, Gullapalli, P.K, Ohtani, K, Akimitsu, K, Izumori, K, Kamitori, S. | | Deposit date: | 2018-03-07 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | X-ray structure of Arthrobacter globiformis M30 ketose 3-epimerase for the production of D-allulose from D-fructose.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
3A9L
 
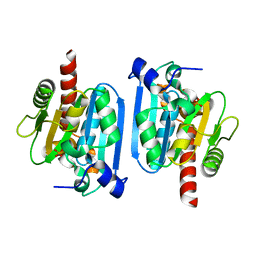 | | Structure of Bacteriophage poly-gamma-glutamate hydrolase | | Descriptor: | PHOSPHATE ION, Poly-gamma-glutamate hydrolase, ZINC ION | | Authors: | Fujimoto, Z, Kimura, K. | | Deposit date: | 2009-10-30 | | Release date: | 2010-11-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of bacteriophage PhiNIT1 zinc peptidase PghP that hydrolyzes gamma-glutamyl linkage of bacterial poly-gamma-glutamate
Proteins, 80, 2012
|
|
5ZQT
 
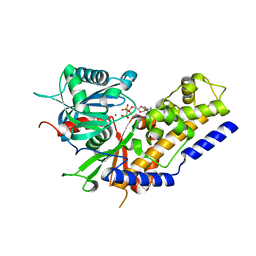 | | Crystal structure of Oryza sativa hexokinase 6 | | Descriptor: | Hexokinase-6, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Matsudaira, K, Mochizuki, S, Yoshida, H, Kamitori, S, Akimitsu, K. | | Deposit date: | 2018-04-20 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal structure of Oryza sativa hexokinase 6
To Be Published
|
|
