6LV1
 
 | | Co- Carbonic Anhydrase II pH 7.8 0 atm CO2 | | Descriptor: | BICARBONATE ION, COBALT (II) ION, Carbonic anhydrase 2, ... | | Authors: | Kim, C.U, Kim, J.K. | | Deposit date: | 2020-02-02 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Elucidating the role of metal ions in carbonic anhydrase catalysis.
Nat Commun, 11, 2020
|
|
6LVA
 
 | | Cu- Carbonic Anhydrase II pH 7.8 20 atm CO2 | | Descriptor: | CARBON DIOXIDE, COPPER (II) ION, Carbonic anhydrase 2, ... | | Authors: | Kim, C.U, Kim, J.K. | | Deposit date: | 2020-02-02 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Elucidating the role of metal ions in carbonic anhydrase catalysis.
Nat Commun, 11, 2020
|
|
6LV3
 
 | | Co- Carbonic Anhydrase II pH 11.0 0 atm CO2 | | Descriptor: | COBALT (II) ION, Carbonic anhydrase 2, GLYCEROL | | Authors: | Kim, C.U, Kim, J.K. | | Deposit date: | 2020-02-02 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Elucidating the role of metal ions in carbonic anhydrase catalysis.
Nat Commun, 11, 2020
|
|
6LV7
 
 | | Ni- Carbonic Anhydrase II pH 11.0 0 atm CO2 | | Descriptor: | Carbonic anhydrase 2, GLYCEROL, NICKEL (II) ION | | Authors: | Kim, C.U, Kim, J.K. | | Deposit date: | 2020-02-02 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Elucidating the role of metal ions in carbonic anhydrase catalysis.
Nat Commun, 11, 2020
|
|
6KM6
 
 | | Human Carbonic Anhydrase II native 15 atm CO2 | | Descriptor: | CARBON DIOXIDE, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Kim, C.U, Kim, J.K. | | Deposit date: | 2019-07-30 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural insights into the effect of active-site mutation on the catalytic mechanism of carbonic anhydrase.
Iucrj, 7, 2020
|
|
6KM2
 
 | | Human Carbonic Anhydrase II V143I variant 15 atm CO2 | | Descriptor: | BICARBONATE ION, CARBON DIOXIDE, Carbonic anhydrase 2, ... | | Authors: | Kim, C.U, Kim, J.K. | | Deposit date: | 2019-07-30 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Structural insights into the effect of active-site mutation on the catalytic mechanism of carbonic anhydrase.
Iucrj, 7, 2020
|
|
3E5N
 
 | | Crystal structure of D-alanine-D-alanine ligase from Xanthomonas oryzae pv. oryzae KACC10331 | | Descriptor: | D-alanine-D-alanine ligase A | | Authors: | Doan, T.N.T, Kim, J.K, Kim, H.S, Ahn, Y.J, Kim, J.G, Lee, B.M, Kang, L.W. | | Deposit date: | 2008-08-14 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of D-alanine-D-alanine ligase from Xanthomonas oryzae pv. oryzae KACC10331
To be published
|
|
3E6G
 
 | | Crystal structure of XometC, a cystathionine c-lyase-like protein from Xanthomonas oryzae pv.oryzae | | Descriptor: | Cystathionine gamma-lyase-like protein | | Authors: | Ngo, H.P.T, Kim, J.K, Kim, H.S, Jung, J.H, Ahn, Y.J, Kim, J.G, Lee, B.M, Kang, H.W, Kang, L.W. | | Deposit date: | 2008-08-15 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of XometC, a cystathionine c-lyase-like protein from Xanthomonas oryzae pv.oryzae
To be published
|
|
7L18
 
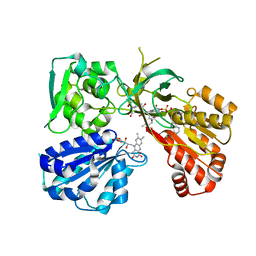 | | Crystal structure of a tandem deletion mutant of rat NADPH-cytochrome P450 reductase | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Hubbard, P.A, Xia, C, Shen, A.L, Kim, J.J.K. | | Deposit date: | 2020-12-14 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.542 Å) | | Cite: | Structural and kinetic investigations of the carboxy terminus of NADPH-cytochrome P450 oxidoreductase.
Arch.Biochem.Biophys., 701, 2021
|
|
3JTR
 
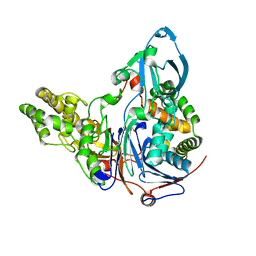 | | Mutations in Cephalosporin Acylase Affecting Stability and Autoproteolysis | | Descriptor: | GLYCEROL, Glutaryl 7-aminocephalosporanic acid acylase | | Authors: | Cho, K.J, Kim, J.K, Lee, J.H, Shin, H.J, Park, S.S, Kim, K.H. | | Deposit date: | 2009-09-14 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural features of cephalosporin acylase reveal the basis of autocatalytic activation.
Biochem.Biophys.Res.Commun., 390, 2009
|
|
3JTQ
 
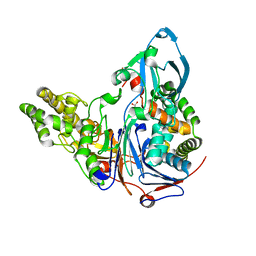 | | Mutations in Cephalosporin Acylase Affecting Stability and Autoproteolysis | | Descriptor: | GLYCEROL, Glutaryl 7-aminocephalosporanic acid acylase | | Authors: | Cho, K.J, Kim, J.K, Lee, J.H, Shin, H.J, Park, S.S, Kim, K.H. | | Deposit date: | 2009-09-14 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural features of cephalosporin acylase reveal the basis of autocatalytic activation.
Biochem.Biophys.Res.Commun., 390, 2009
|
|
6NJR
 
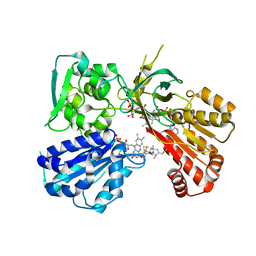 | | Spin-Labeled T177C/A637C Mutant of Rat CYPOR | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Xia, C, Kim, J.J.K. | | Deposit date: | 2019-01-04 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Functional Studies of the Membrane-Binding Domain of NADPH-Cytochrome P450 Oxidoreductase.
Biochemistry, 58, 2019
|
|
5FA6
 
 | | wild type human CYPOR | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Xia, C, Marohnic, C, Panda, S, Masters, B.S, Kim, J.J.K. | | Deposit date: | 2015-12-11 | | Release date: | 2016-08-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Instability of the Human Cytochrome P450 Reductase A287P Variant Is the Major Contributor to Its Antley-Bixler Syndrome-like Phenotype.
J.Biol.Chem., 291, 2016
|
|
5EMN
 
 | | Crystal Structure of Human NADPH-Cytochrome P450 Reductase(A287P mutant) | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Xia, C, Marohnic, C, Panda, S, Masters, B.S, Kim, J.J.K. | | Deposit date: | 2015-11-06 | | Release date: | 2016-08-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Instability of the Human Cytochrome P450 Reductase A287P Variant Is the Major Contributor to Its Antley-Bixler Syndrome-like Phenotype.
J.Biol.Chem., 291, 2016
|
|
4IXZ
 
 | | Native structure of cystathionine gamma lyase (XometC) from xanthomonas oryzae pv. oryzae at pH 9.0 | | Descriptor: | BETA-MERCAPTOETHANOL, BICARBONATE ION, Cystathionine gamma-lyase-like protein, ... | | Authors: | Ngo, H.P.T, Kim, J.K, Kang, L.W. | | Deposit date: | 2013-01-28 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | PLP undergoes conformational changes during the course of an enzymatic reaction.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4IYO
 
 | | Crystal structure of cystathionine gamma lyase from Xanthomonas oryzae pv. oryzae (XometC) in complex with E-site serine, A-site serine, A-site external aldimine structure with aminoacrylate and A-site iminopropionate intermediates | | Descriptor: | 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, AMINO-ACRYLATE, Cystathionine gamma-lyase-like protein, ... | | Authors: | Ngo, H.P.T, Kim, J.K, Kang, L.W. | | Deposit date: | 2013-01-29 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | PLP undergoes conformational changes during the course of an enzymatic reaction.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4IY7
 
 | | crystal structure of cystathionine gamma lyase (XometC) from Xanthomonas oryzae pv. oryzae in complex with E-site serine, A-site external aldimine structure with serine and A-site external aldimine structure with aminoacrylate intermediates | | Descriptor: | (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-serine, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, Cystathionine gamma-lyase-like protein, ... | | Authors: | Ngo, H.P.T, Kim, J.K, Kang, L.W. | | Deposit date: | 2013-01-28 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | PLP undergoes conformational changes during the course of an enzymatic reaction.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4NFX
 
 | |
4IXS
 
 | | Native structure of xometc at ph 5.2 | | Descriptor: | CARBONATE ION, Cystathionine gamma-lyase-like protein, GLYCEROL | | Authors: | Ngo, H.P.T, Kim, J.K, Kang, L.W. | | Deposit date: | 2013-01-28 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | PLP undergoes conformational changes during the course of an enzymatic reaction.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4NFW
 
 | | Structure and atypical hydrolysis mechanism of the Nudix hydrolase Orf153 (YmfB) from Escherichia coli | | Descriptor: | MANGANESE (II) ION, Putative Nudix hydrolase ymfB, SULFATE ION | | Authors: | Hong, M.K, Kim, J.K, Kang, L.W. | | Deposit date: | 2013-11-01 | | Release date: | 2014-05-14 | | Last modified: | 2015-03-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Divalent metal ion-based catalytic mechanism of the Nudix hydrolase Orf153 (YmfB) from Escherichia coli
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3HE8
 
 | | Structural study of Clostridium thermocellum Ribose-5-Phosphate Isomerase B | | Descriptor: | GLYCEROL, Ribose-5-phosphate isomerase | | Authors: | Kang, L.W, Kim, J.K, Jung, J.H, Hong, M.K. | | Deposit date: | 2009-05-08 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Clostridium thermocellum ribose-5-phosphate isomerase B reveals properties critical for fast enzyme kinetics.
Appl.Microbiol.Biotechnol., 90, 2011
|
|
3HEE
 
 | | Structural study of Clostridium thermocellum Ribose-5-Phosphate Isomerase B and ribose-5-phosphate | | Descriptor: | RIBOSE-5-PHOSPHATE, Ribose-5-phosphate isomerase | | Authors: | Kang, L.W, Kim, J.K, Jung, J.H, Hong, M.K. | | Deposit date: | 2009-05-08 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Clostridium thermocellum ribose-5-phosphate isomerase B reveals properties critical for fast enzyme kinetics.
Appl.Microbiol.Biotechnol., 90, 2011
|
|
3DKU
 
 | | Crystal structure of Nudix hydrolase Orf153, ymfB, from Escherichia coli K-1 | | Descriptor: | Putative phosphohydrolase | | Authors: | Hong, M.K, Kim, J.K, Jung, J.H, Jung, J.W, Choi, J.Y, Kang, L.W. | | Deposit date: | 2008-06-26 | | Release date: | 2009-06-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal structure of Nudix hydrolase Orf153, ymfB, from Escherichia coli K-1.
To be Published
|
|
3HR4
 
 | | Human iNOS Reductase and Calmodulin Complex | | Descriptor: | CALCIUM ION, Calmodulin, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Xia, C, Misra, I, Iyanaki, T, Kim, J.J.K. | | Deposit date: | 2009-06-08 | | Release date: | 2009-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Regulation of Interdomain Interactions by CaM in Inducible Nitric Oxide Synthase
J.Biol.Chem., 2009
|
|
3PH3
 
 | | Clostridium thermocellum Ribose-5-Phosphate Isomerase B with d-ribose | | Descriptor: | D-ribose, Ribose-5-phosphate isomerase | | Authors: | Jung, J, Kim, J.K, Yeom, S.J, Ahn, Y.J, Oh, D.K, Kang, L.W. | | Deposit date: | 2010-11-03 | | Release date: | 2011-06-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of Clostridium thermocellum ribose-5-phosphate isomerase B reveals properties critical for fast enzyme kinetics.
Appl.Microbiol.Biotechnol., 90, 2011
|
|
