1L8O
 
 | | Molecular basis for the local conformational rearrangement of human phosphoserine phosphatase | | Descriptor: | L-3-phosphoserine phosphatase, PHOSPHATE ION, SERINE | | Authors: | Kim, H.Y, Heo, Y.S, Kim, J.H, Park, M.H, Moon, J, Park, S.Y, Lee, T.G, Jeon, Y.H, Ro, S, Hwang, K.Y. | | Deposit date: | 2002-03-21 | | Release date: | 2003-04-01 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis for the local conformational rearrangement of human phosphoserine phosphatase
J.Biol.Chem., 277, 2002
|
|
1L8L
 
 | | Molecular basis for the local confomational rearrangement of human phosphoserine phosphatase | | Descriptor: | D-2-AMINO-3-PHOSPHONO-PROPIONIC ACID, L-3-phosphoserine phosphatase | | Authors: | Kim, H.Y, Heo, Y.S, Kim, J.H, Park, M.H, Moon, J, Park, S.Y, Lee, T.G, Jeon, Y.H, Ro, S, Hwang, K.Y. | | Deposit date: | 2002-03-21 | | Release date: | 2003-04-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Molecular basis for the local conformational rearrangement of human phosphoserine phosphatase.
J.Biol.Chem., 277, 2002
|
|
1AD6
 
 | | DOMAIN A OF HUMAN RETINOBLASTOMA TUMOR SUPPRESSOR | | Descriptor: | RETINOBLASTOMA TUMOR SUPPRESSOR | | Authors: | Kim, H.Y, Cho, Y. | | Deposit date: | 1997-02-21 | | Release date: | 1998-08-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural similarity between the pocket region of retinoblastoma tumour suppressor and the cyclin-box.
Nat.Struct.Biol., 4, 1997
|
|
1GH6
 
 | | RETINOBLASTOMA POCKET COMPLEXED WITH SV40 LARGE T ANTIGEN | | Descriptor: | Large T antigen, Retinoblastoma-associated protein | | Authors: | Kim, H.Y, Cho, Y. | | Deposit date: | 2000-11-15 | | Release date: | 2001-11-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the inactivation of retinoblastoma tumor suppressor by SV40 large T antigen.
EMBO J., 20, 2001
|
|
4G4W
 
 | | Crystal structure of peptidoglycan-associated lipoprotein from Acinetobacter baumannii | | Descriptor: | ALANINE, GLYCEROL, Peptidoglycan-associated lipoprotein, ... | | Authors: | Lee, W.C, Song, J.H, Park, J.S, Kim, H.Y. | | Deposit date: | 2012-07-16 | | Release date: | 2013-07-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Enantiomer-dependent amino acid binding affinity of OmpA-like domains from Acinetobacter baumannii peptidoglycan-associated lipoprotein and OmpA
to be published
|
|
2KV1
 
 | | Insights into Function, Catalytic Mechanism and Fold Evolution of Mouse Selenoprotein Methionine Sulfoxide Reductase B1 through Structural Analysis | | Descriptor: | Methionine-R-sulfoxide reductase B1, ZINC ION | | Authors: | Aachmann, F.L, Sal, L.S, Kim, H.Y, Gladyshev, V.N, Dikiy, A. | | Deposit date: | 2010-03-04 | | Release date: | 2010-03-16 | | Last modified: | 2014-04-09 | | Method: | SOLUTION NMR | | Cite: | Insights into function, catalytic mechanism, and fold evolution of selenoprotein methionine sulfoxide reductase B1 through structural analysis
J.Biol.Chem., 285, 2010
|
|
5G54
 
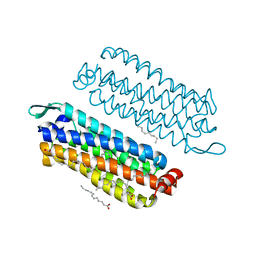 | | The crystal structure of light-driven chloride pump ClR at pH 4.5 | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Kim, K.L, Kwon, S.K, Jun, S.H, Cha, J.S, Kim, H.Y, Kim, J.H, Cho, H.S. | | Deposit date: | 2016-05-19 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure and Functional Characterization of a Light-Driven Chloride Pump Having an Ntq Motif.
Nat.Commun., 7, 2016
|
|
5G28
 
 | | The crystal structure of light-driven chloride pump ClR at pH 6.0. | | Descriptor: | CHLORIDE ION, CHLORIDE PUMPING RHODOPSIN, OLEIC ACID, ... | | Authors: | Kim, K.L, Kwon, S.K, Jun, S.H, Cha, J.S, Kim, H.Y, Kim, J.H, Cho, H.S. | | Deposit date: | 2016-04-07 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal Structure and Functional Characterization of a Light-Driven Chloride Pump Having an Ntq Motif.
Nat.Commun., 7, 2016
|
|
5G2D
 
 | | The crystal structure of light-driven chloride pump ClR (T102N) mutant at pH 4.5. | | Descriptor: | CHLORIDE ION, CHLORIDE PUMP RHODOPSIN, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, K.L, Kwon, S.K, Jun, S.H, Cha, J.S, Kim, H.Y, Kim, J.H, Cho, H.S. | | Deposit date: | 2016-04-07 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure and Functional Characterization of a Light-Driven Chloride Pump Having an Ntq Motif.
Nat.Commun., 7, 2016
|
|
5J4F
 
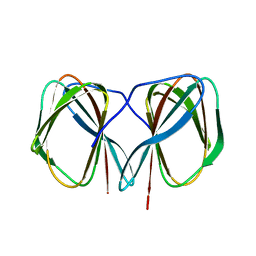 | | Crystal structure of the N-terminally His6-tagged HP0902, an uncharacterized protein from Helicobacter pylori 26695 | | Descriptor: | Uncharacterized protein | | Authors: | Sim, D.-W, Lee, W.-C, Kim, H.Y, Kim, J.-H, Won, H.-S. | | Deposit date: | 2016-04-01 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural identification of the lipopolysaccharide-binding capability of a cupin-family protein from Helicobacter pylori
FEBS Lett., 590, 2016
|
|
5J4G
 
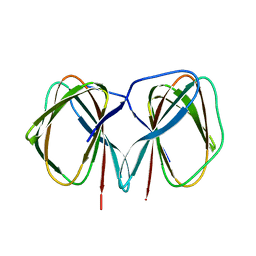 | | Crystal structure of the C-terminally His6-tagged HP0902, an uncharacterized protein from Helicobacter pylori 26695 | | Descriptor: | Uncharacterized protein | | Authors: | Sim, D.W, Lee, W.C, Kim, H.Y, Kim, J.H, Won, H.S. | | Deposit date: | 2016-04-01 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural identification of the lipopolysaccharide-binding capability of a cupin-family protein from Helicobacter pylori
FEBS Lett., 590, 2016
|
|
5G2C
 
 | | The crystal structure of light-driven chloride pump ClR (T102D) mutant at pH 4.5. | | Descriptor: | CHLORIDE ION, CHLORIDE PUMPING RHODOPSIN, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, K.L, Kwon, S.K, Jun, S.H, Cha, J.S, Kim, H.Y, Kim, J.H, Cho, H.S. | | Deposit date: | 2016-04-07 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal Structure and Functional Characterization of a Light-Driven Chloride Pump Having an Ntq Motif.
Nat.Commun., 7, 2016
|
|
5G2A
 
 | | The crystal structure of light-driven chloride pump ClR at pH 6.0 with Bromide ion. | | Descriptor: | BROMIDE ION, CHLORIDE PUMPING RHODOPSIN, RETINAL | | Authors: | Kim, K.L, Kwon, S.K, Jun, S.H, Cha, J.S, Kim, H.Y, Kim, J.H, Cho, H.S. | | Deposit date: | 2016-04-07 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal Structure and Functional Characterization of a Light-Driven Chloride Pump Having an Ntq Motif.
Nat.Commun., 7, 2016
|
|
3NR1
 
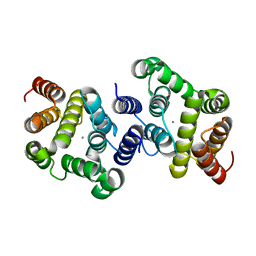 | | A metazoan ortholog of SpoT hydrolyzes ppGpp and plays a role in starvation responses | | Descriptor: | HD domain-containing protein 3, MANGANESE (II) ION | | Authors: | Sun, D.W, Kim, H.Y, Kim, K.J, Jeon, Y.H, Chung, J. | | Deposit date: | 2010-06-30 | | Release date: | 2010-09-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A metazoan ortholog of SpoT hydrolyzes ppGpp and functions in starvation responses
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NQW
 
 | | A metazoan ortholog of SpoT hydrolyzes ppGpp and plays a role in starvation responses | | Descriptor: | CG11900, MANGANESE (II) ION, SULFATE ION | | Authors: | Sun, D.W, Kim, H.Y, Kim, K.J, Jeon, Y.H, Chung, J. | | Deposit date: | 2010-06-30 | | Release date: | 2010-09-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A metazoan ortholog of SpoT hydrolyzes ppGpp and functions in starvation responses
Nat.Struct.Mol.Biol., 17, 2010
|
|
3INA
 
 | | Crystal structure of heparin lyase I H151A mutant complexed with a dodecasaccharide heparin | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-3,6-di-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, CALCIUM ION, Heparin lyase I | | Authors: | Han, Y.H, Ryu, K.S, Kim, H.Y, Jeon, Y.H. | | Deposit date: | 2009-08-12 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural snapshots of heparin depolymerization by heparin lyase I
J.Biol.Chem., 284, 2009
|
|
3IN9
 
 | | Crystal structure of heparin lyase I complexed with disaccharide heparin | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CALCIUM ION, Heparin lyase I | | Authors: | Han, Y.H, Ryu, K.S, Kim, H.Y, Jeon, Y.H. | | Deposit date: | 2009-08-12 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural snapshots of heparin depolymerization by heparin lyase I
J.Biol.Chem., 284, 2009
|
|
5FJG
 
 | | The crystal structure of light-driven chloride pump ClR in pH 4.5. | | Descriptor: | ANHYDRORETINOL, CHLORIDE ION, CHLORIDE PUMPING RHODOPSIN, ... | | Authors: | Kim, K.L, Kwon, S.K, Jun, S.H, Cha, J.S, Kim, H.Y, Kim, J.H, Cho, H.S. | | Deposit date: | 2015-10-07 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Light-Driven Chloride Pump Clr in Ph 4.5.
To be Published
|
|
4G4V
 
 | |
4G4Y
 
 | |
4G4X
 
 | | Crystal structure of peptidoglycan-associated lipoprotein from Acinetobacter baumannii | | Descriptor: | D-ALANINE, GLYCEROL, Peptidoglycan-associated lipoprotein, ... | | Authors: | Lee, W.C, Song, J.H, Park, J.S, Kim, H.Y. | | Deposit date: | 2012-07-16 | | Release date: | 2013-07-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Enantiomer-dependent amino acid binding affinity of OmpA-like domains from Acinetobacter baumannii peptidoglycan-associated lipoprotein and OmpA
To be Published
|
|
4G88
 
 | | Crystal structure of OmpA peptidoglycan-binding domain from Acinetobacter baumannii | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, Outer membrane protein Omp38, S,R MESO-TARTARIC ACID | | Authors: | Lee, W.C, Song, J.H, Park, J.S, Kim, H.Y. | | Deposit date: | 2012-07-22 | | Release date: | 2013-07-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Enantiomer-dependent amino acid binding affinity of OmpA-like domains from Acinetobacter baumannii peptidoglycan-associated lipoprotein and OmpA
to be published
|
|
4RCP
 
 | | Crystal structure of Plk1 Polo-box domain in complex with PL-2 | | Descriptor: | 1,2-ETHANEDIOL, PL-2, Serine/threonine-protein kinase PLK1 | | Authors: | Lee, W.C, Song, J.H, Kim, H.Y. | | Deposit date: | 2014-09-16 | | Release date: | 2014-11-12 | | Last modified: | 2015-01-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A new class of peptidomimetics targeting the polo-box domain of polo-like kinase 1.
J.Med.Chem., 58, 2015
|
|
4G4Z
 
 | |
4MMN
 
 | | Structural and biochemical analysis of type II free methionine-R-sulfoxide reductase from Thermoplasma acidophilum | | Descriptor: | Putative uncharacterized protein Ta0848 | | Authors: | Kim, H.S, Kwak, G.H, Lee, K.T, Jo, C.H, Hwang, K.Y, Kim, H.Y. | | Deposit date: | 2013-09-09 | | Release date: | 2014-08-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and biochemical analysis of a type II free methionine-R-sulfoxide reductase from Thermoplasma acidophilum
Arch.Biochem.Biophys., 560, 2014
|
|
