4Q6Q
 
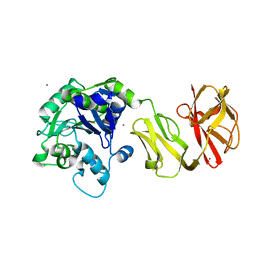 | | Structural analysis of the Zn-form II of Helicobacter pylori Csd4, a D,L-carboxypeptidase | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, CALCIUM ION, Conserved hypothetical secreted protein, ... | | Authors: | Kim, H.S, Kim, J, Im, H.N, An, D.R, Lee, M, Hesek, D, Mobashery, S, Kim, J.Y, Cho, K, Yoon, H.J, Han, B.W, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-04-23 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the recognition of muramyltripeptide by Helicobacter pylori Csd4, a D,L-carboxypeptidase controlling the helical cell shape
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4Q6N
 
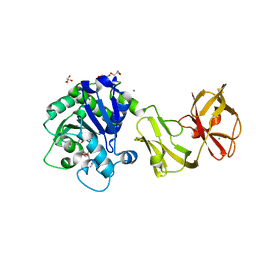 | | Structural analysis of the tripeptide-bound form of Helicobacter pylori Csd4, a D,L-carboxypeptidase | | Descriptor: | CALCIUM ION, Conserved hypothetical secreted protein, GLYCEROL, ... | | Authors: | Kim, H.S, Kim, J, Im, H.N, An, D.R, Lee, M, Hesek, D, Mobashery, S, Kim, J.Y, Cho, K, Yoon, H.J, Han, B.W, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-04-23 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for the recognition of muramyltripeptide by Helicobacter pylori Csd4, a D,L-carboxypeptidase controlling the helical cell shape
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4Q6M
 
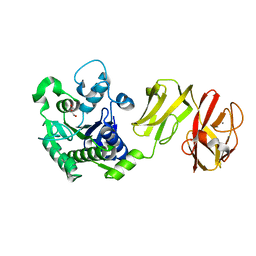 | | Structural analysis of the apo-form of Helicobacter pylori Csd4, a D,L-carboxypeptidase | | Descriptor: | CALCIUM ION, Conserved hypothetical secreted protein, GLYCEROL | | Authors: | Kim, H.S, Kim, J, Im, H.N, An, D.R, Lee, M, Hesek, D, Mobashery, S, Kim, J.Y, Cho, K, Yoon, H.J, Han, B.W, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-04-23 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of muramyltripeptide by Helicobacter pylori Csd4, a D,L-carboxypeptidase controlling the helical cell shape
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4Q6O
 
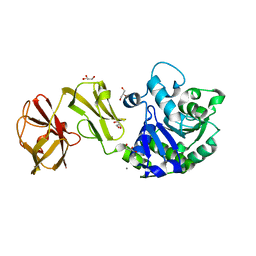 | | Structural analysis of the mDAP-bound form of Helicobacter pylori Csd4, a D,L-carboxypeptidase | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, CALCIUM ION, Conserved hypothetical secreted protein, ... | | Authors: | Kim, H.S, Kim, J, Im, H.N, An, D.R, Lee, M, Hesek, D, Mobashery, S, Kim, J.Y, Cho, K, Yoon, H.J, Han, B.W, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-04-23 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural basis for the recognition of muramyltripeptide by Helicobacter pylori Csd4, a D,L-carboxypeptidase controlling the helical cell shape
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4Q6P
 
 | | Structural analysis of the Zn-form I of Helicobacter pylori Csd4, a D,L-carboxypeptidase | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, CALCIUM ION, Conserved hypothetical secreted protein, ... | | Authors: | Kim, H.S, Kim, J, Im, H.N, An, D.R, Lee, M, Hesek, D, Mobashery, S, Kim, J.Y, Cho, K, Yoon, H.J, Han, B.W, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-04-23 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural basis for the recognition of muramyltripeptide by Helicobacter pylori Csd4, a D,L-carboxypeptidase controlling the helical cell shape
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3QY6
 
 | | Crystal structures of YwqE from Bacillus subtilis and CpsB from Streptococcus pneumoniae, unique metal-dependent tyrosine phosphatases | | Descriptor: | FE (III) ION, MAGNESIUM ION, Tyrosine-protein phosphatase YwqE | | Authors: | Kim, H.S, Lee, S.J, Yoon, H.J, An, D.R, Kim, D.J, Kim, S.-J, Suh, S.W. | | Deposit date: | 2011-03-03 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of YwqE from Bacillus subtilis and CpsB from Streptococcus pneumoniae, unique metal-dependent tyrosine phosphatases.
J.Struct.Biol., 175, 2011
|
|
3QY8
 
 | | Crystal structures of YwqE from Bacillus subtilis and CpsB from Streptococcus pneumoniae, unique metal-dependent tyrosine phosphatases | | Descriptor: | FE (III) ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kim, H.S, Lee, S.J, Yoon, H.J, An, D.R, Kim, D.J, Kim, S.-J, Suh, S.W. | | Deposit date: | 2011-03-03 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of YwqE from Bacillus subtilis and CpsB from Streptococcus pneumoniae, unique metal-dependent tyrosine phosphatases.
J.Struct.Biol., 175, 2011
|
|
3QY7
 
 | | Crystal structures of YwqE from Bacillus subtilis and CpsB from Streptococcus pneumoniae, unique metal-dependent tyrosine phosphatases | | Descriptor: | FE (III) ION, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Kim, H.S, Lee, S.J, Yoon, H.J, An, D.R, Kim, D.J, Kim, S.-J, Suh, S.W. | | Deposit date: | 2011-03-03 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structures of YwqE from Bacillus subtilis and CpsB from Streptococcus pneumoniae, unique metal-dependent tyrosine phosphatases.
J.Struct.Biol., 175, 2011
|
|
6IOZ
 
 | | Structural insights of idursulfase beta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kim, H, Kim, D, Hong, J, Lee, K, Seo, J, Oh, B.H. | | Deposit date: | 2018-11-01 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural insights of idursulfase beta
To Be Published
|
|
7WRQ
 
 | | Structure of Human IGF1/IGFBP3/ALS Ternary Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin-like growth factor-binding protein 3, Insulin-like growth factor-binding protein complex acid labile subunit, ... | | Authors: | Kim, H, Fu, Y, Kim, H.M. | | Deposit date: | 2022-01-27 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for assembly and disassembly of the IGF/IGFBP/ALS ternary complex
Nat Commun, 13, 2022
|
|
4MMN
 
 | | Structural and biochemical analysis of type II free methionine-R-sulfoxide reductase from Thermoplasma acidophilum | | Descriptor: | Putative uncharacterized protein Ta0848 | | Authors: | Kim, H.S, Kwak, G.H, Lee, K.T, Jo, C.H, Hwang, K.Y, Kim, H.Y. | | Deposit date: | 2013-09-09 | | Release date: | 2014-08-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and biochemical analysis of a type II free methionine-R-sulfoxide reductase from Thermoplasma acidophilum
Arch.Biochem.Biophys., 560, 2014
|
|
4MN7
 
 | | Structural and biochemical analysis of type II free methionine-R-sulfoxide reductase from Thermoplasma acidophilum | | Descriptor: | METHIONINE SULFOXIDE, Putative uncharacterized protein Ta0848 | | Authors: | Kim, H.S, Kwak, G.H, Lee, K.T, Jo, C.H, Hwang, K.Y, Kim, H.Y. | | Deposit date: | 2013-09-10 | | Release date: | 2014-08-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and biochemical analysis of a type II free methionine-R-sulfoxide reductase from Thermoplasma acidophilum
Arch.Biochem.Biophys., 560, 2014
|
|
1A7K
 
 | | GLYCOSOMAL GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE IN A MONOCLINIC CRYSTAL FORM | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Kim, H, Hol, W.G.J. | | Deposit date: | 1998-03-16 | | Release date: | 1998-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Leishmania mexicana glycosomal glyceraldehyde-3-phosphate dehydrogenase in a new crystal form confirms the putative physiological active site structure.
J.Mol.Biol., 278, 1998
|
|
7CPA
 
 | |
4DXN
 
 | |
4DXM
 
 | |
4DXO
 
 | |
7KWA
 
 | | Structure of DCN1 bound to N-((4S,5S)-3-(aminomethyl)-7-ethyl-4-(4-fluorophenyl)-6-oxo-1-phenyl-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl)-3-(trifluoromethyl)benzamide | | Descriptor: | Endolysin,DCN1-like protein 1, N-[(4S,5S)-3-(aminomethyl)-7-ethyl-4-(4-fluorophenyl)-6-oxo-1-phenyl-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl]-3-(trifluoromethyl)benzamide | | Authors: | Kim, H.S, Hammill, J.T, Schulman, B.A, Guy, R.K, Scott, D.C. | | Deposit date: | 2020-11-30 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.572 Å) | | Cite: | Improvement of Oral Bioavailability of Pyrazolo-Pyridone Inhibitors of the Interaction of DCN1/2 and UBE2M.
J.Med.Chem., 64, 2021
|
|
4DXQ
 
 | |
4DXI
 
 | |
4DXP
 
 | |
1A5C
 
 | | FRUCTOSE-1,6-BISPHOSPHATE ALDOLASE FROM PLASMODIUM FALCIPARUM | | Descriptor: | FRUCTOSE-1,6-BISPHOSPHATE ALDOLASE | | Authors: | Kim, H, Certa, U, Dobeli, H, Jakob, P, Hol, W.G.J. | | Deposit date: | 1998-02-13 | | Release date: | 1998-06-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of fructose-1,6-bisphosphate aldolase from the human malaria parasite Plasmodium falciparum.
Biochemistry, 37, 1998
|
|
1BPN
 
 | |
1BLL
 
 | |
1BPM
 
 | |
