6R4T
 
 | |
6R5E
 
 | |
6R4U
 
 | |
6RB4
 
 | |
6R5D
 
 | |
6R4S
 
 | |
4BPU
 
 | | Crystal structure of human primase in heterodimeric form, comprising PriS and truncated PriL lacking the C-terminal Fe-S domain. | | Descriptor: | DNA PRIMASE LARGE SUBUNIT, DNA PRIMASE SMALL SUBUNIT, GLYCEROL, ... | | Authors: | Kilkenny, M.L, Perera, R.L, Pellegrini, L. | | Deposit date: | 2013-05-28 | | Release date: | 2013-09-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of Human Primase Reveal Design of Nucleotide Elongation Site and Mode of Pol Alpha Tethering
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4BPX
 
 | | Crystal structure of human primase in complex with the primase- binding motif of DNA polymerase alpha | | Descriptor: | DNA POLYMERASE ALPHA CATALYTIC SUBUNIT, DNA PRIMASE LARGE SUBUNIT, DNA PRIMASE SMALL SUBUNIT, ... | | Authors: | Kilkenny, M.L, Perera, R.L, Pellegrini, L. | | Deposit date: | 2013-05-28 | | Release date: | 2013-09-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structures of Human Primase Reveal Design of Nucleotide Elongation Site and Mode of Pol Alpha Tethering
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4BPW
 
 | | Crystal structure of human primase bound to UTP | | Descriptor: | DNA PRIMASE LARGE SUBUNIT, DNA PRIMASE SMALL SUBUNIT, MAGNESIUM ION, ... | | Authors: | Kilkenny, M.L, Perera, R.L, Pellegrini, L. | | Deposit date: | 2013-05-28 | | Release date: | 2013-09-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Structures of Human Primase Reveal Design of Nucleotide Elongation Site and Mode of Pol Alpha Tethering
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2VXC
 
 | |
2VXB
 
 | | Structure of the Crb2-BRCT2 domain | | Descriptor: | DNA REPAIR PROTEIN RHP9, PRASEODYMIUM ION | | Authors: | Kilkenny, M.L, Roe, S.M, Pearl, L.H. | | Deposit date: | 2008-07-03 | | Release date: | 2008-08-12 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Functional Analysis of the Crb2-Brct2 Domain Reveals Distinct Roles in Checkpoint Signaling and DNA Damage Repair.
Genes Dev., 22, 2008
|
|
4WWM
 
 | | X-ray crystal structure of Sulfolobus solfataricus Urm1 | | Descriptor: | SULFATE ION, Uncharacterized protein | | Authors: | Bray, S.M, Anjum, S.R, Blackwood, J.K, Kilkenny, M.L, Coelho, M.A, Foster, B.M, Pellegrini, L, Robinson, N.P. | | Deposit date: | 2014-11-11 | | Release date: | 2015-09-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Involvement of a eukaryotic-like ubiquitin-related modifier in the proteasome pathway of the archaeon Sulfolobus acidocaldarius.
Nat Commun, 6, 2015
|
|
3TM7
 
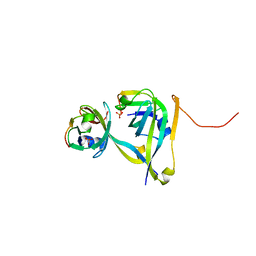 | | Processed Aspartate Decarboxylase Mutant with Asn72 mutated to Ala | | Descriptor: | Aspartate 1-decarboxylase alpha chain, Aspartate 1-decarboxylase beta chain, SULFATE ION | | Authors: | Webb, M.E, Lobley, C.M.C, Soliman, F, Kilkenny, M.L, Smith, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2011-08-31 | | Release date: | 2012-04-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Escherichia coli aspartate alpha-decarboxylase Asn72Ala: probing the role of Asn72 in pyruvoyl cofactor formation
Acta Crystallogr.,Sect.F, 68, 2012
|
|
5MQI
 
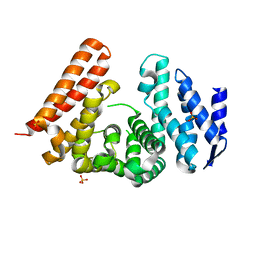 | |
6TAZ
 
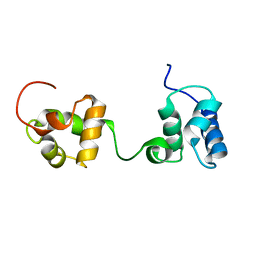 | | Timeless couples G quadruplex detection with processing by DDX11 during DNA replication | | Descriptor: | Protein timeless homolog | | Authors: | Lerner Koch, L, Holzer, S, Kilkenny, M.L, Murat, P, Svikovic, S, Schiavone, D, Bittleston, A, Maman, J.D, Branzei, D, Stott, K, Pellegrini, L, Sale, E.J. | | Deposit date: | 2019-10-31 | | Release date: | 2020-07-01 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Timeless couples G-quadruplex detection with processing by DDX11 helicase during DNA replication.
Embo J., 39, 2020
|
|
3G65
 
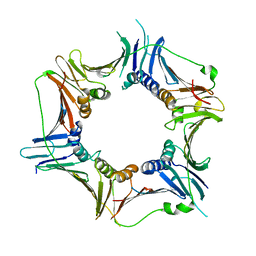 | | Crystal Structure of the Human Rad9-Rad1-Hus1 DNA Damage Checkpoint Complex | | Descriptor: | Cell cycle checkpoint control protein RAD9A, Cell cycle checkpoint protein RAD1, Checkpoint protein HUS1 | | Authors: | Dore, A.S, Kilkenny, M.L, Rzechorzek, N.J, Pearl, L.H. | | Deposit date: | 2009-02-06 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the rad9-rad1-hus1 DNA damage checkpoint complex--implications for clamp loading and regulation.
Mol.Cell, 34, 2009
|
|
4BA9
 
 | | The structural basis for the coordination of Y-family Translesion DNA Polymerases by Rev1 | | Descriptor: | DNA POLYMERASE KAPPA, DNA REPAIR PROTEIN REV1, MAGNESIUM ION, ... | | Authors: | Grummitt, C.G, Kilkenny, M.L, Frey, A, Roe, S.M, Oliver, A.W, Sale, J.E, Pearl, L.H. | | Deposit date: | 2012-09-12 | | Release date: | 2013-09-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | The Structural Basis for the Coordination of Y- Family Translesion DNA Polymerases by Rev1
To be Published
|
|
4B08
 
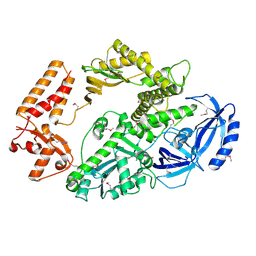 | | Yeast DNA polymerase alpha, Selenomethionine protein | | Descriptor: | DNA POLYMERASE ALPHA CATALYTIC SUBUNIT A | | Authors: | Perera, R.L, Torella, R, Klinge, S, Kilkenny, M.L, Maman, J.D, Pellegrini, L. | | Deposit date: | 2012-06-29 | | Release date: | 2013-02-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Mechanism for Priming DNA Synthesis by Yeast DNA Polymerase Alpha
Elife, 2, 2013
|
|
4AZD
 
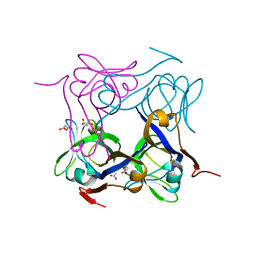 | | T57V mutant of aspartate decarboxylase | | Descriptor: | ASPARTATE 1-DECARBOXYLASE, MALONATE ION | | Authors: | Webb, M.E, Yorke, B.A, Kershaw, T, Lovelock, S, Lobley, C.M.C, Kilkenny, M.L, Smith, A.G, Blundell, T.L, Pearson, A.R, Abell, C. | | Deposit date: | 2012-06-25 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Threonine 57 is Required for the Post-Translational Activation of Escherichia Coli Aspartate Alpha-Decarboxylase
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2IZO
 
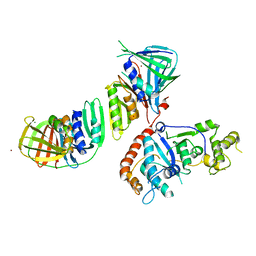 | | Structure of an Archaeal PCNA1-PCNA2-FEN1 Complex | | Descriptor: | DNA POLYMERASE SLIDING CLAMP B, DNA POLYMERASE SLIDING CLAMP C, FLAP STRUCTURE-SPECIFIC ENDONUCLEASE, ... | | Authors: | Dore, A.S, Kilkenny, M.L, Roe, S.M, Pearl, L.H. | | Deposit date: | 2006-07-25 | | Release date: | 2006-09-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of an Archaeal PCNA1-PCNA2-Fen1 Complex: Elucidating PCNA Subunit and Client Enzyme Specificity.
Nucleic Acids Res., 34, 2006
|
|
1PQF
 
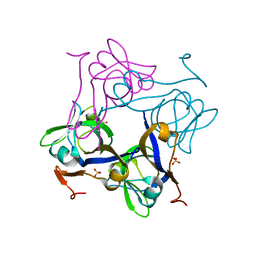 | | Glycine 24 to Serine mutation of aspartate decarboxylase | | Descriptor: | Aspartate 1-decarboxylase, SULFATE ION | | Authors: | Schmitzberger, F, Kilkenny, M.L, Lobley, C.M.C, Webb, M.E, Vinkovic, M, Matak-Vinkovic, D, Witty, M, Chirgadze, D.Y, Smith, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2003-06-18 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural constraints on protein self-processing in L-aspartate-alpha-decarboxylase
Embo J., 22, 2003
|
|
1PYQ
 
 | | Unprocessed Aspartate Decarboxylase Mutant, with Alanine inserted at position 24 | | Descriptor: | Aspartate 1-decarboxylase, SULFATE ION | | Authors: | Schmitzberger, F, Kilkenny, M.L, Lobley, C.M.C, Webb, M.E, Vinkovic, M, Matak-Vinkovic, D, Witty, M, Chirgadze, D.Y, Smith, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2003-07-09 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Constraints on protein self-processing in L-aspartate-alpha-decarboxylase
Embo J., 22, 2003
|
|
1PPY
 
 | | Native precursor of pyruvoyl dependent Aspartate decarboxylase | | Descriptor: | Aspartate 1-decarboxylase precursor, SULFATE ION | | Authors: | Schmitzberger, F, Kilkenny, M.L, Lobley, C.M.C, Webb, M.E, Vinkovic, M, Matak-Vinkovic, D, Witty, M, Chirgadze, D.Y, Smith, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2003-06-17 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural constraints on protein self-processing in L-aspartate-alpha-decarboxylase
Embo J., 22, 2003
|
|
1PYU
 
 | | Processed Aspartate Decarboxylase Mutant with Ser25 mutated to Cys | | Descriptor: | Aspartate 1-decarboxylase alfa chain, Aspartate 1-decarboxylase beta chain, SULFATE ION | | Authors: | Schmitzberger, F, Kilkenny, M.L, Lobley, C.M.C, Webb, M.E, Vinkovic, M, Matak-Vinkovic, D, Witty, M, Chirgadze, D.Y, Smith, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2003-07-09 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Constraints on protein self-processing in L-aspartate-alpha-decarboxylase
Embo J., 22, 2003
|
|
1PQE
 
 | | S25A mutant of pyruvoyl dependent aspartate decarboxylase | | Descriptor: | Aspartate 1-decarboxylase | | Authors: | Schmitzberger, F, Kilkenny, M.L, Lobley, C.M.C, Webb, M.E, Vinkovic, M, Matak-Vinkovic, D, Witty, M, Chirgadze, D.Y, Smith, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2003-06-18 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural constraints on protein self-processing in L-aspartate-alpha-decarboxylase
Embo J., 22, 2003
|
|
