7U0W
 
 | |
5TSG
 
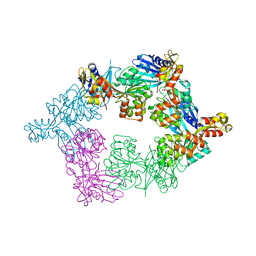 | | PilB from Geobacter metallireducens bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | McCallum, M, Tammam, S, Khan, A, Burrows, L, Howell, P.L. | | Deposit date: | 2016-10-28 | | Release date: | 2017-05-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.4011 Å) | | Cite: | The molecular mechanism of the type IVa pilus motors.
Nat Commun, 8, 2017
|
|
6S5I
 
 | |
6RIR
 
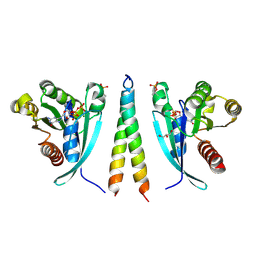 | | Crystal structure of phosphorylated Rab8a in complex with the Rab-binding domain of RILPL2 | | Descriptor: | GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Waschbusch, D, Purlyte, E, Alessi, D.R, Khan, A.R. | | Deposit date: | 2019-04-25 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.767 Å) | | Cite: | Structural Basis for Rab8a Recruitment of RILPL2 via LRRK2 Phosphorylation of Switch 2.
Structure, 28, 2020
|
|
2K36
 
 | | Structure ensemble Backbone and side-chain 1H, 13C, and 15N Chemical Shift Assignments, 1H-15N RDCs and 1H-1H nOe restraints for protein K7 from the Vaccinia virus | | Descriptor: | Protein K7 | | Authors: | Kalverda, A.P, Thompson, G.S, Vogel, A, Schr der, M, Bowie, A.G, Khan, A.R, Homans, S.W. | | Deposit date: | 2008-04-22 | | Release date: | 2008-10-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Poxvirus K7 protein adopts a Bcl-2 fold: biochemical mapping of its interactions with human DEAD box RNA helicase DDX3.
J.Mol.Biol., 385, 2009
|
|
7Q5C
 
 | | Crystal structure of OmpG in space group 96 | | Descriptor: | Outer membrane porin G, SODIUM ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Nguyen, T.T.M, Khan, A.R, Barringer, R, McManus, J.J, Race, P.R. | | Deposit date: | 2021-11-03 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.717 Å) | | Cite: | Experimental phase diagrams to optimise OmpG
To Be Published
|
|
8GII
 
 | | TEM-1 Beta Lactamase Variant 80.a | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, TEM-1 Variant 80.a | | Authors: | Fram, B.F, Gauthier, N.P, Khan, A.R, Sander, C. | | Deposit date: | 2023-03-14 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Simultaneous enhancement of multiple functional properties using evolution-informed protein design.
Nat Commun, 15, 2024
|
|
6HDU
 
 | |
5TSH
 
 | | PilB from Geobacter metallireducens bound to AMP-PNP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | McCallum, M, Tammam, S, Khan, A, Burrows, L, Howell, P.L. | | Deposit date: | 2016-10-28 | | Release date: | 2017-05-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The molecular mechanism of the type IVa pilus motors.
Nat Commun, 8, 2017
|
|
6S8X
 
 | | Crystal structure of the Rab-binding domain of FIP2 | | Descriptor: | HEXANE-1,6-DIOL, Rab11 family-interacting protein 2 | | Authors: | Kearney, A.M, Khan, A.R. | | Deposit date: | 2019-07-10 | | Release date: | 2020-08-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of the Rab-binding domain of Rab11 family-interacting protein 2.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6EYW
 
 | |
6EZL
 
 | |
2OA7
 
 | |
2OAC
 
 | |
6WHE
 
 | |
2OAD
 
 | |
2YFQ
 
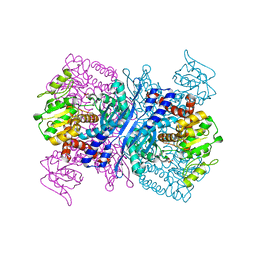 | | Crystal structure of Glutamate dehydrogenase from Peptoniphilus asaccharolyticus | | Descriptor: | NAD-SPECIFIC GLUTAMATE DEHYDROGENASE, SULFATE ION | | Authors: | Oliveira, T, Panjikar, S, Carrigan, J.B, Sharkey, M.A, Hamza, M, Engel, P.C, Khan, A.R. | | Deposit date: | 2011-04-07 | | Release date: | 2011-11-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Crystal Structure of Nad-Dependent Peptoniphilus Asaccharolyticus Glutamate Dehydrogenase Reveals Determinants of Cofactor Specificity.
J.Struct.Biol., 177, 2012
|
|
2W88
 
 | |
2XSJ
 
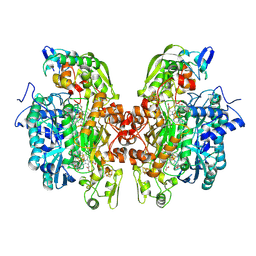 | | Structure of desulforubidin from Desulfomicrobium norvegicum | | Descriptor: | IRON/SULFUR CLUSTER, SIROHEME, SULFITE ION, ... | | Authors: | Oliveira, T.F, Khan, A.R, Pereira, I.A.C, Archer, M. | | Deposit date: | 2010-09-29 | | Release date: | 2011-08-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights Into Dissimilatory Sulfite Reductases: Structure of Desulforubidin from Desulfomicrobium Norvegicum
Front.Microbiol., 2, 2011
|
|
2YFH
 
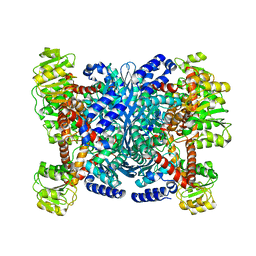 | | Structure of a Chimeric Glutamate Dehydrogenase | | Descriptor: | GLUTAMATE DEHYDROGENASE, NAD-SPECIFIC GLUTAMATE DEHYDROGENASE | | Authors: | Oliveira, T, Panjikar, S, Sharkey, M.A, Carrigan, J.B, Hamza, M, Engel, P.C, Khan, A.R. | | Deposit date: | 2011-04-05 | | Release date: | 2012-04-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.695 Å) | | Cite: | Crystal Structure of a Chimaeric Bacterial Glutamate Dehydrogenase.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
2W8C
 
 | |
1BXQ
 
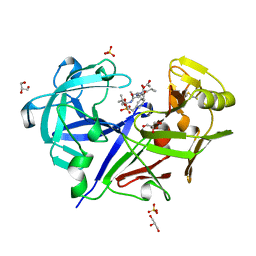 | | ACID PROTEINASE (PENICILLOPEPSIN) COMPLEX WITH PHOSPHONATE INHIBITOR. | | Descriptor: | 2-[(1R)-1-(N-(3-METHYLBUTANOYL)-L-VALYL-L-ASPARAGINYL)-AMINO)-3-METHYLBUTYL]HYDROXYPHOSPHINYLOXY]-3-PHENYLPROPANOIC ACID METHYLESTER, ACETATE ION, GLYCEROL, ... | | Authors: | Parrish, J.C, Khan, A.R, Fraser, M.E, Smith, W.W, Bartlett, P.A, James, M.N.G. | | Deposit date: | 1998-10-07 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Lowering the entropic barrier for binding conformationally flexible inhibitors to enzymes.
Biochemistry, 37, 1998
|
|
6ETA
 
 | |
4D0G
 
 | | Structure of Rab14 in complex with Rab-Coupling Protein (RCP) | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RAB11 FAMILY-INTERACTING PROTEIN 1, ... | | Authors: | Lall, P, Khan, A.R. | | Deposit date: | 2014-04-25 | | Release date: | 2015-05-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Function Analyses of the Interactions between Rab11 and Rab14 Small Gtpases with Their Shared Effector Rab Coupling Protein (Rcp).
J.Biol.Chem., 290, 2015
|
|
3TYI
 
 | | Crystal Structure of Cytochrome c - p-Sulfonatocalix[4]arene Complexes | | Descriptor: | 25,26,27,28-tetrahydroxypentacyclo[19.3.1.1~3,7~.1~9,13~.1~15,19~]octacosa-1(25),3(28),4,6,9(27),10,12,15(26),16,18,21,23-dodecaene-5,11,17,23-tetrasulfonic acid, Cytochrome c iso-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mc Govern, R.E, Fernandes, H, Khan, A.R, Crowley, P.B. | | Deposit date: | 2011-09-26 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Protein camouflage in cytochrome c-calixarene complexes.
Nat Chem, 4, 2012
|
|
