3ALU
 
 | | Crystal structure of CEL-IV complexed with Raffinose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Lectin CEL-IV, ... | | Authors: | Hatakeyama, T, Hozawa, T, Ishii, K, Kamiya, T, Goda, S, Kusunoki, M, Unno, H. | | Deposit date: | 2010-08-07 | | Release date: | 2011-01-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Galactose recognition by a tetrameric C-type lectin, CEL-IV, containing the EPN carbohydrate recognition motif
J.Biol.Chem., 286, 2011
|
|
3ALT
 
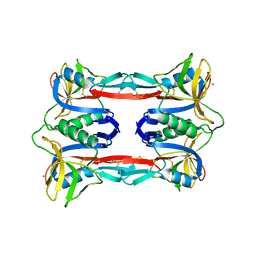 | | Crystal structure of CEL-IV complexed with Melibiose | | Descriptor: | CALCIUM ION, Lectin CEL-IV, C-type, ... | | Authors: | Hatakeyama, T, Hozawa, T, Ishii, K, Kamiya, T, Goda, S, Kusunoki, M, Unno, H. | | Deposit date: | 2010-08-07 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Galactose recognition by a tetrameric C-type lectin, CEL-IV, containing the EPN carbohydrate recognition motif
J.Biol.Chem., 286, 2011
|
|
3ALS
 
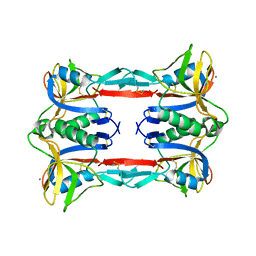 | | Crystal structure of CEL-IV | | Descriptor: | CALCIUM ION, Lectin CEL-IV, C-type | | Authors: | Hatakeyama, T, Hozawa, T, Ishii, K, Kamiya, T, Goda, S, Kusunoki, M, Unno, H. | | Deposit date: | 2010-08-07 | | Release date: | 2011-01-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Galactose recognition by a tetrameric C-type lectin, CEL-IV, containing the EPN carbohydrate recognition motif
J.Biol.Chem., 286, 2011
|
|
8DCC
 
 | |
4HJ7
 
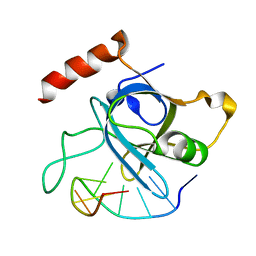 | |
4HIK
 
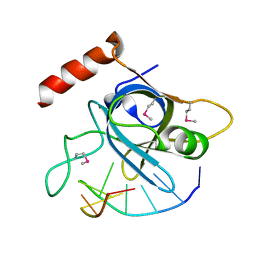 | |
4HIO
 
 | |
4HJ9
 
 | |
4HIM
 
 | |
2N9L
 
 | | 1H, 13C, and 15N Chemical Shift Assignments for in-cell GB1 | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Ikeya, T, Hanashima, T, Hosoya, S, Shimazaki, M, Ikeda, S, Mishima, M, Guentert, P, Ito, Y. | | Deposit date: | 2015-11-30 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Improved in-cell structure determination of proteins at near-physiological concentration
Sci Rep, 6, 2016
|
|
2N9K
 
 | | 1H, 13C, and 15N Chemical Shift Assignments for in vitro GB1 | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Ikeya, T, Hanashima, T, Hosoya, S, Shimazaki, M, Ikeda, S, Mishima, M, Guentert, P, Ito, Y. | | Deposit date: | 2015-11-26 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Improved in-cell structure determination of proteins at near-physiological concentration
Sci Rep, 6, 2016
|
|
4HJ5
 
 | |
4HJ8
 
 | |
4HID
 
 | |
4HJA
 
 | |
3JZ4
 
 | | Crystal structure of E. coli NADP dependent enzyme | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Succinate-semialdehyde dehydrogenase [NADP+] | | Authors: | Langendorf, C.G, Key, T.L.G, Fenalti, G, Kan, W.T, Buckle, A.M, Caradoc-Davies, T, Tuck, K.L, Law, R.H.P, Whisstock, J.C. | | Deposit date: | 2009-09-22 | | Release date: | 2010-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The X-ray crystal structure of Escherichia coli succinic semialdehyde dehydrogenase; structural insights into NADP+/enzyme interactions.
Plos One, 5, 2010
|
|
3VP6
 
 | | Structural characterization of Glutamic Acid Decarboxylase; insights into the mechanism of autoinactivation | | Descriptor: | 4-oxo-4H-pyran-2,6-dicarboxylic acid, GLYCEROL, Glutamate decarboxylase 1 | | Authors: | Langendorf, C.G, Tuck, K.L, Key, T.L.G, Rosado, C.J, Wong, A.S.M, Fenalti, G, Buckle, A.M, Law, R.H.P, Whisstock, J.C. | | Deposit date: | 2012-02-27 | | Release date: | 2013-01-16 | | Last modified: | 2013-08-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of the mechanism through which human glutamic acid decarboxylase auto-activates
Biosci.Rep., 33, 2013
|
|
2LX0
 
 | |
6K4I
 
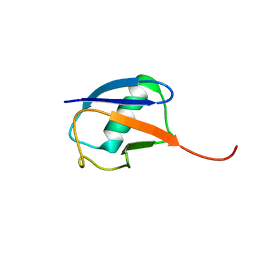 | | The partially disordered conformation of ubiquitin (Q41N variant) | | Descriptor: | ubiquitin | | Authors: | Wakamoto, T, Ikeya, T, Kitazawa, S, Baxter, N.J, Williamson, M.P, Kitahara, R. | | Deposit date: | 2019-05-24 | | Release date: | 2019-10-30 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Paramagnetic relaxation enhancement-assisted structural characterization of a partially disordered conformation of ubiquitin.
Protein Sci., 28, 2019
|
|
3N35
 
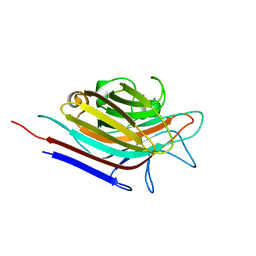 | | Erythrina corallodendron lectin mutant (Y106G) with N-Acetylgalactosamine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, CALCIUM ION, Lectin, ... | | Authors: | Thamotharan, S, Karthikeyan, T, Kulkarni, K.A, Shetty, K.N, Surolia, A, Vijayan, M, Suguna, K. | | Deposit date: | 2010-05-19 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Modification of the sugar specificity of a plant lectin: structural studies on a point mutant of Erythrina corallodendron lectin.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
6TLB
 
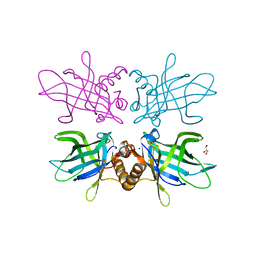 | | Plasmodium falciparum lipocalin (PF3D7_0925900) | | Descriptor: | GLYCEROL, SODIUM ION, Serine/threonine protein kinase | | Authors: | Burda, P.C, Crosskey, T.D, Lauk, K, Wilmanns, M, Gilberger, T.W. | | Deposit date: | 2019-12-02 | | Release date: | 2020-06-24 | | Last modified: | 2024-11-06 | | Method: | SOLUTION SCATTERING (2.85 Å), X-RAY DIFFRACTION | | Cite: | Structure-Based Identification and Functional Characterization of a Lipocalin in the Malaria Parasite Plasmodium falciparum.
Cell Rep, 31, 2020
|
|
8AU4
 
 | | Structural insights reveal a heterotetramer between oncogenic K-Ras4BG12V and Rgl2, a RalA/B activator | | Descriptor: | Ral guanine nucleotide dissociation stimulator-like 2 | | Authors: | Tariq, M, Ikeya, T, Togashi, N, Fairall, L, Alejo, C.B, Kamei, S, Alonso, B.R, Campillo, M.A.M, Hudson, A, Ito, Y, Schwabe, J, Dominguez, C, Tanaka, K. | | Deposit date: | 2022-08-25 | | Release date: | 2023-08-23 | | Last modified: | 2023-10-25 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the complex of oncogenic KRas4B G12V and Rgl2, a RalA/B activator.
Life Sci Alliance, 7, 2024
|
|
7EN4
 
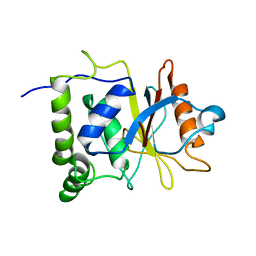 | | Multi-state structure determination and dynamics analysis elucidate a new ubiquitin-recognition mechanism of yeast ubiquitin C-terminal hydrolase. | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase YUH1 | | Authors: | Okada, M, Tateishi, Y, Nojiri, E, Mikawa, T, Rajesh, S, Ogasawa, H, Ueda, T, Yagi, H, Kohno, T, Kigawa, T, Shimada, I, Guentert, P, Yutaka, I, Ikeya, T. | | Deposit date: | 2021-04-15 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Multi-state structure determination and dynamics analysis elucidate a new ubiquitin-recognition mechanism of yeast ubiquitin C-terminal hydrolase.
To Be Published
|
|
7Y4N
 
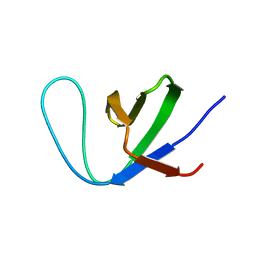 | | Insight into the C-terminal SH3 domain mediated binding of Drosophila Drk to Sos and Dos | | Descriptor: | Growth factor receptor-bound protein 2 | | Authors: | Pooppadi, M.S, Ikeya, T, Sugasawa, H, Watanabe, R, Mishima, M, Inomata, K, Ito, Y. | | Deposit date: | 2022-06-15 | | Release date: | 2022-08-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Insight into the C-terminal SH3 domain mediated binding of Drosophila Drk to Sos and Dos.
Biochem.Biophys.Res.Commun., 625, 2022
|
|
5H4S
 
 | |
