4CDL
 
 | | Crystal Structure of Retro-aldolase RA110.4-6 Complexed with Inhibitor 1-(6-methoxy-2-naphthalenyl)-1,3-butanedione | | 分子名称: | (2E)-1-(6-methoxynaphthalen-2-yl)but-2-en-1-one, STEROID DELTA-ISOMERASE | | 著者 | Pinkas, D.M, Studer, S, Obexer, R, Giger, L, Gruetter, M.G, Baker, D, Hilvert, D. | | 登録日 | 2013-11-01 | | 公開日 | 2014-11-12 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Active Site Plasticity of a Computationally Designed Retro-Aldolase Enzyme
To be Published
|
|
1K51
 
 | | A G55A Mutation Induces 3D Domain Swapping in the B1 Domain of Protein L from Peptostreptococcus magnus | | 分子名称: | Protein L, ZINC ION | | 著者 | O'Neill, J.W, Kim, D.E, Johnsen, K, Baker, D, Zhang, K.Y.J. | | 登録日 | 2001-10-09 | | 公開日 | 2001-12-05 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Single-site mutations induce 3D domain swapping in the B1 domain of protein L from Peptostreptococcus magnus.
Structure, 9, 2001
|
|
1K52
 
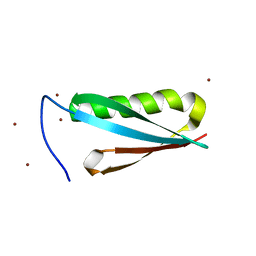 | | Monomeric Protein L B1 Domain with a K54G mutation | | 分子名称: | Protein L, ZINC ION | | 著者 | O'Neill, J.W, Kim, D.E, Johnsen, K, Baker, D, Zhang, K.Y.J. | | 登録日 | 2001-10-09 | | 公開日 | 2001-12-05 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Single-site mutations induce 3D domain swapping in the B1 domain of protein L from Peptostreptococcus magnus.
Structure, 9, 2001
|
|
3U13
 
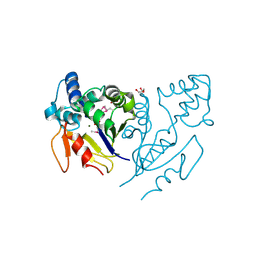 | | Crystal Structure of de Novo design of cystein esterase ECH13 at the resolution 1.6A, Northeast Structural Genomics Consortium Target OR51 | | 分子名称: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | Kuzin, A, Su, M, Seetharaman, J, Sahdev, S, Xiao, R, Kohan, E, Richter, F, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2011-09-29 | | 公開日 | 2011-11-23 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Computational design of catalytic dyads and oxyanion holes for ester hydrolysis.
J.Am.Chem.Soc., 134, 2012
|
|
1KH0
 
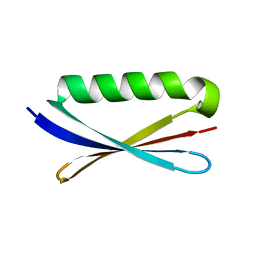 | | Accurate Computer Base Design of a New Backbone Conformation in the Second Turn of Protein L | | 分子名称: | protein L | | 著者 | O'Neill, J.W, Kuhlman, B, Kim, D.E, Zhang, K.Y, Baker, D. | | 登録日 | 2001-11-28 | | 公開日 | 2002-01-23 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Accurate computer-based design of a new backbone conformation in the second turn of protein L.
J.Mol.Biol., 315, 2002
|
|
3KFU
 
 | | Crystal structure of the transamidosome | | 分子名称: | Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit B, Glutamyl-tRNA(Gln) amidotransferase subunit A, Glutamyl-tRNA(Gln) amidotransferase subunit C, ... | | 著者 | Blaise, M, Bailly, M, Frechin, M, Thirup, S, Becker, H.D, Kern, D. | | 登録日 | 2009-10-28 | | 公開日 | 2010-08-25 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structure of a transfer-ribonucleoprotein particle that promotes asparagine formation.
Embo J., 29, 2010
|
|
3U1O
 
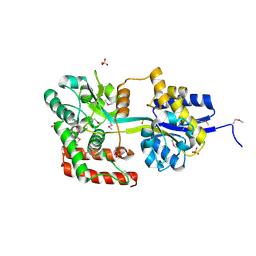 | | THREE DIMENSIONAL STRUCTURE OF DE NOVO DESIGNED CYSTEINE ESTERASE ECH19, Northeast Structural Genomics Consortium Target OR49 | | 分子名称: | De Novo design cysteine esterase ECH19, SODIUM ION, SULFATE ION | | 著者 | Kuzin, A, Su, M, Lew, S, Forouhar, F, Seetharaman, J, Daya, P, Xiao, R, Ciccosanti, C, Richter, F, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2011-09-30 | | 公開日 | 2011-10-26 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.494 Å) | | 主引用文献 | Computational design of catalytic dyads and oxyanion holes for ester hydrolysis.
J.Am.Chem.Soc., 134, 2012
|
|
3U1V
 
 | | X-ray Structure of De Novo design cysteine esterase FR29, Northeast Structural Genomics Consortium Target OR52 | | 分子名称: | De Novo design cysteine esterase FR29 | | 著者 | Kuzin, A, Su, M, Vorobiev, S.M, Seetharaman, J, Patel, D, Xiao, R, Ciccosanti, C, Richter, F, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2011-09-30 | | 公開日 | 2011-12-07 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.797 Å) | | 主引用文献 | Computational design of catalytic dyads and oxyanion holes for ester hydrolysis.
J.Am.Chem.Soc., 134, 2012
|
|
1UJZ
 
 | | Crystal structure of the E7_C/Im7_C complex; a computationally designed interface between the colicin E7 DNase and the Im7 Immunity protein | | 分子名称: | Designed Colicin E7 DNase, Designed Colicin E7 immunity protein | | 著者 | Kortemme, T, Joachimiak, L.A, Bullock, A.N, Schuler, A.D, Stoddard, B.L, Baker, D. | | 登録日 | 2003-08-13 | | 公開日 | 2004-04-06 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Computational redesign of protein-protein interaction specificity
NAT.STRUCT.MOL.BIOL., 11, 2004
|
|
8SKD
 
 | |
8SKX
 
 | |
1MI0
 
 | | Crystal Structure of the redesigned protein G variant NuG2 | | 分子名称: | immunoglobulin-binding protein G | | 著者 | Nauli, S, Kuhlman, B, Le Trong, I, Stenkamp, R.E, Teller, D.C, Baker, D. | | 登録日 | 2002-08-21 | | 公開日 | 2002-09-18 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal structures and increased stabilization of the protein G variants with switched folding pathways NuG1 and NuG2
Biochemistry, 11, 2002
|
|
8SKE
 
 | |
3TC7
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR62. | | 分子名称: | ACETIC ACID, Indole-3-glycerol phosphate synthase, PHOSPHATE ION | | 著者 | Vorobiev, S, Su, M, Bjelic, S, Kipnis, Y, Wang, L, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2011-08-08 | | 公開日 | 2011-08-24 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Exploration of alternate catalytic mechanisms and optimization strategies for retroaldolase design.
J.Mol.Biol., 426, 2014
|
|
3GEQ
 
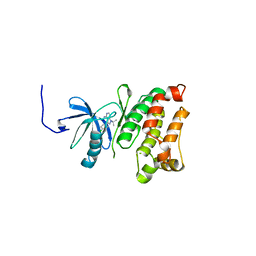 | | Structural basis for the chemical rescue of Src kinase activity | | 分子名称: | 1-TERT-BUTYL-3-(4-CHLORO-PHENYL)-1H-PYRAZOLO[3,4-D]PYRIMIDIN-4-YLAMINE, Proto-oncogene tyrosine-protein kinase Src | | 著者 | Muratore, K.E, Seeliger, M.A, Wang, Z, Fomina, D, Neiswinger, J, Havranek, J.J, Baker, D, Kuriyan, J, Cole, P.A. | | 登録日 | 2009-02-25 | | 公開日 | 2009-04-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Comparative analysis of mutant tyrosine kinase chemical rescue.
Biochemistry, 48, 2009
|
|
6BES
 
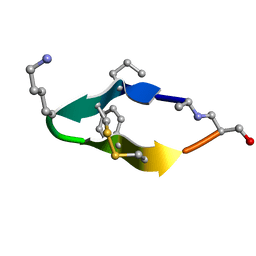 | | Solution structure of de novo macrocycle design11_ss | | 分子名称: | (DAL)Q(DPR)(DCY)(DLY)DS(DTY)(DCY)P(DSN) | | 著者 | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, D. | | 登録日 | 2017-10-25 | | 公開日 | 2017-12-27 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
6BEW
 
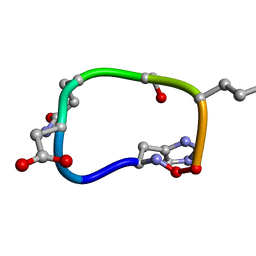 | | Solution structure of de novo macrocycle design7.2 | | 分子名称: | (DHI)P(DAS)(DGN)(DSN)(DGL)P | | 著者 | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, D. | | 登録日 | 2017-10-25 | | 公開日 | 2017-12-27 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
6BET
 
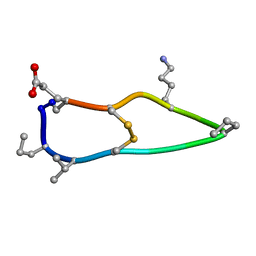 | | Solution structure of de novo macrocycle design12_ss | | 分子名称: | H(DPR)(DVA)CIP(DPR)E(DLY)VC(DGL) | | 著者 | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, D. | | 登録日 | 2017-10-25 | | 公開日 | 2017-12-27 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
6BF5
 
 | | Solution structure of de novo macrocycle design7.3a | | 分子名称: | QDP(DPR)K(DTH)(DAS) | | 著者 | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, D. | | 登録日 | 2017-10-25 | | 公開日 | 2018-01-10 | | 最終更新日 | 2023-11-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
6BEO
 
 | | Solution structure of de novo macrocycle design9.1 | | 分子名称: | (DPR)PY(DHI)PKDL(DGN) | | 著者 | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, D. | | 登録日 | 2017-10-25 | | 公開日 | 2017-12-27 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
6BER
 
 | | Solution structure of de novo macrocycle design10.2 | | 分子名称: | E(DVA)DP(DGL)(DHI)(DPR)N(DAL)(DPR) | | 著者 | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, D. | | 登録日 | 2017-10-25 | | 公開日 | 2017-12-27 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
6BEQ
 
 | | Solution structure of de novo macrocycle design10.1 | | 分子名称: | AAR(DVA)(DPR)R(DLE)(DTH)PE | | 著者 | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, D. | | 登録日 | 2017-10-25 | | 公開日 | 2018-01-10 | | 最終更新日 | 2023-11-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
6BEU
 
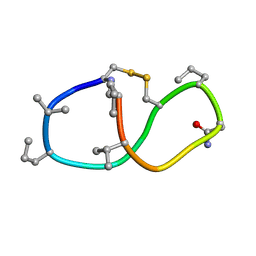 | | Solution structure of de novo macrocycle design14_ss | | 分子名称: | (DCY)N(DVA)(DPR)DVYC(DPR)(DSG)KY(DVA)(DPR) | | 著者 | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, D. | | 登録日 | 2017-10-25 | | 公開日 | 2017-12-27 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
1MHX
 
 | | Crystal Structures of the redesigned protein G variant NuG1 | | 分子名称: | immunoglobulin-binding protein G | | 著者 | Nauli, S, Kuhlman, B, Le Trong, I, Stenkamp, R.E, Teller, D.C, Baker, D. | | 登録日 | 2002-08-21 | | 公開日 | 2002-09-18 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structures and increased stabilization of the protein G variants with switched folding pathways NuG1 and NuG2
Protein Sci., 11, 2002
|
|
6BF3
 
 | | Solution structure of de novo macrocycle design7.3a | | 分子名称: | QDP(DPR)K(2TL)(DAS) | | 著者 | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, D. | | 登録日 | 2017-10-25 | | 公開日 | 2018-01-10 | | 最終更新日 | 2023-11-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
