6X9Z
 
 | |
5V2O
 
 | | De Novo Design of Novel Covalent Constrained Meso-size Peptide Scaffolds with Unique Tertiary Structures | | 分子名称: | 1,3,5-BENZENETRICARBOXYLIC ACID, GLYCEROL, NONAETHYLENE GLYCOL, ... | | 著者 | Dang, B, Wu, H, Mulligan, V.K, Mravic, M, Wu, Y, Lemmin, T, Ford, A, Silva, D, Baker, D, DeGrado, W.F. | | 登録日 | 2017-03-06 | | 公開日 | 2017-10-04 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | De novo design of covalently constrained mesosize protein scaffolds with unique tertiary structures.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5V2G
 
 | | De Novo Design of Novel Covalent Constrained Meso-size Peptide Scaffolds with Unique Tertiary Structures | | 分子名称: | 1,3,5-tris(bromomethyl)benzene, 20-mer Peptide | | 著者 | Dang, B, Wu, H, Mulligan, V.K, Mravic, M, Wu, Y, Lemmin, T, Ford, A, Silva, D, Baker, D, DeGrado, W.F. | | 登録日 | 2017-03-03 | | 公開日 | 2017-09-27 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | De novo design of covalently constrained mesosize protein scaffolds with unique tertiary structures.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6BF3
 
 | | Solution structure of de novo macrocycle design7.3a | | 分子名称: | QDP(DPR)K(2TL)(DAS) | | 著者 | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, D. | | 登録日 | 2017-10-25 | | 公開日 | 2018-01-10 | | 最終更新日 | 2023-11-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
6BE7
 
 | | Solution structure of de novo macrocycle Design8.1 | | 分子名称: | DDPT(DPR)(DAR)Q(DGN) | | 著者 | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, D. | | 登録日 | 2017-10-24 | | 公開日 | 2018-01-03 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
6BES
 
 | | Solution structure of de novo macrocycle design11_ss | | 分子名称: | (DAL)Q(DPR)(DCY)(DLY)DS(DTY)(DCY)P(DSN) | | 著者 | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, D. | | 登録日 | 2017-10-25 | | 公開日 | 2017-12-27 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
4A29
 
 | | Structure of the engineered retro-aldolase RA95.0 | | 分子名称: | 1-(6-METHOXYNAPHTHALEN-2-YL)BUTANE-1,3-DIONE, D-MALATE, ENGINEERED RETRO-ALDOL ENZYME RA95.0 | | 著者 | Giger, L, Caner, S, Kast, P, Baker, D, Ban, N, Hilvert, D. | | 登録日 | 2011-09-23 | | 公開日 | 2012-11-07 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Evolution of a designed retro-aldolase leads to complete active site remodeling.
Nat.Chem.Biol., 9, 2013
|
|
4A2R
 
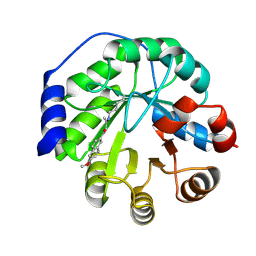 | | Structure of the engineered retro-aldolase RA95.5-5 | | 分子名称: | 1-(6-METHOXYNAPHTHALEN-2-YL)BUTANE-1,3-DIONE, INDOLE-3-GLYCEROL PHOSPHATE SYNTHASE | | 著者 | Giger, L, Caner, S, Kast, P, Baker, D, Ban, N, Hilvert, D. | | 登録日 | 2011-09-28 | | 公開日 | 2012-11-07 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.302 Å) | | 主引用文献 | Evolution of a designed retro-aldolase leads to complete active site remodeling.
Nat.Chem.Biol., 9, 2013
|
|
6BEW
 
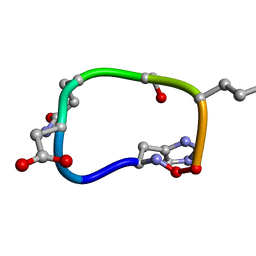 | | Solution structure of de novo macrocycle design7.2 | | 分子名称: | (DHI)P(DAS)(DGN)(DSN)(DGL)P | | 著者 | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, D. | | 登録日 | 2017-10-25 | | 公開日 | 2017-12-27 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
6BET
 
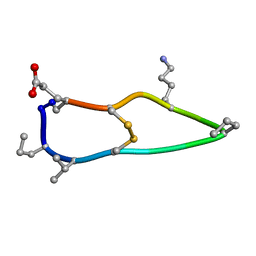 | | Solution structure of de novo macrocycle design12_ss | | 分子名称: | H(DPR)(DVA)CIP(DPR)E(DLY)VC(DGL) | | 著者 | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, D. | | 登録日 | 2017-10-25 | | 公開日 | 2017-12-27 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
6BF5
 
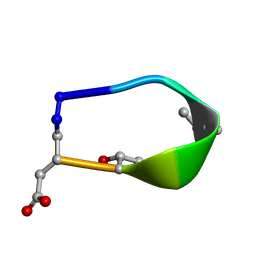 | | Solution structure of de novo macrocycle design7.3a | | 分子名称: | QDP(DPR)K(DTH)(DAS) | | 著者 | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, D. | | 登録日 | 2017-10-25 | | 公開日 | 2018-01-10 | | 最終更新日 | 2023-11-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
6BEO
 
 | | Solution structure of de novo macrocycle design9.1 | | 分子名称: | (DPR)PY(DHI)PKDL(DGN) | | 著者 | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, D. | | 登録日 | 2017-10-25 | | 公開日 | 2017-12-27 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
6BER
 
 | | Solution structure of de novo macrocycle design10.2 | | 分子名称: | E(DVA)DP(DGL)(DHI)(DPR)N(DAL)(DPR) | | 著者 | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, D. | | 登録日 | 2017-10-25 | | 公開日 | 2017-12-27 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
6BEQ
 
 | | Solution structure of de novo macrocycle design10.1 | | 分子名称: | AAR(DVA)(DPR)R(DLE)(DTH)PE | | 著者 | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, D. | | 登録日 | 2017-10-25 | | 公開日 | 2018-01-10 | | 最終更新日 | 2023-11-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
6BEU
 
 | | Solution structure of de novo macrocycle design14_ss | | 分子名称: | (DCY)N(DVA)(DPR)DVYC(DPR)(DSG)KY(DVA)(DPR) | | 著者 | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, D. | | 登録日 | 2017-10-25 | | 公開日 | 2017-12-27 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
8FRE
 
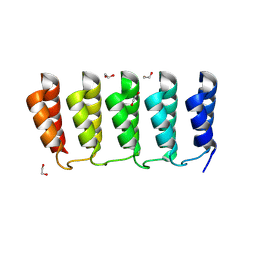 | | Designed loop protein RBL4 | | 分子名称: | 1,2-ETHANEDIOL, RBL4 | | 著者 | Jude, K.M, Jiang, H, Baker, D, Garcia, K.C. | | 登録日 | 2023-01-06 | | 公開日 | 2024-04-17 | | 最終更新日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | De novo design of buttressed loops for sculpting protein functions.
Nat.Chem.Biol., 2024
|
|
8FRF
 
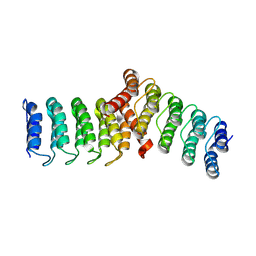 | |
8FNE
 
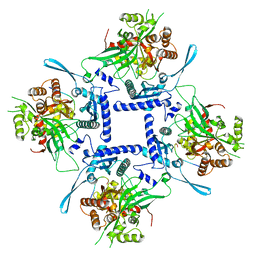 | | phiPA3 PhuN Tetramer, p2 | | 分子名称: | Maltose/maltodextrin-binding periplasmic protein, PhuN | | 著者 | Nieweglowska, E.S, Brilot, A.F, Mendez-Moran, M, Kokontis, C, Baek, M, Li, J, Cheng, Y, Baker, D, Bondy-Denomy, J, Agard, D.A. | | 登録日 | 2022-12-27 | | 公開日 | 2023-03-01 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | The phi PA3 phage nucleus is enclosed by a self-assembling 2D crystalline lattice.
Nat Commun, 14, 2023
|
|
8FV5
 
 | | Representation of 16-mer phiPA3 PhuN Lattice, p2 | | 分子名称: | Maltose/maltodextrin-binding periplasmic protein, phiPA3 PhuN | | 著者 | Nieweglowska, E.S, Brilot, A.F, Mendez-Moran, M, Kokontis, C, Baek, M, Li, J, Cheng, Y, Baker, D, Bondy-Denomy, J, Agard, D.A. | | 登録日 | 2023-01-18 | | 公開日 | 2023-03-01 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (4.21 Å) | | 主引用文献 | The phi PA3 phage nucleus is enclosed by a self-assembling 2D crystalline lattice.
Nat Commun, 14, 2023
|
|
3GEQ
 
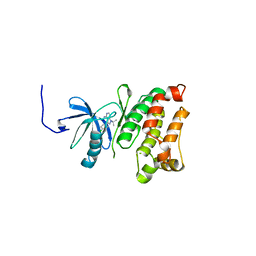 | | Structural basis for the chemical rescue of Src kinase activity | | 分子名称: | 1-TERT-BUTYL-3-(4-CHLORO-PHENYL)-1H-PYRAZOLO[3,4-D]PYRIMIDIN-4-YLAMINE, Proto-oncogene tyrosine-protein kinase Src | | 著者 | Muratore, K.E, Seeliger, M.A, Wang, Z, Fomina, D, Neiswinger, J, Havranek, J.J, Baker, D, Kuriyan, J, Cole, P.A. | | 登録日 | 2009-02-25 | | 公開日 | 2009-04-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Comparative analysis of mutant tyrosine kinase chemical rescue.
Biochemistry, 48, 2009
|
|
6MSP
 
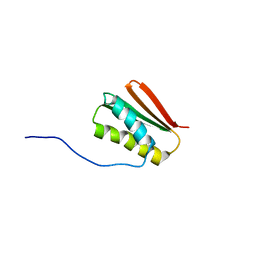 | | De novo Designed Protein Foldit3 | | 分子名称: | De novo Designed Protein Foldit3 | | 著者 | Liu, G, Ishida, Y, Swapna, G.V.T, Kleinfelter, S, Koepnick, B, Baker, D, Montelione, G.T. | | 登録日 | 2018-10-17 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | De novo protein design by citizen scientists.
Nature, 570, 2019
|
|
6MSQ
 
 | | Crystal structure of pRO-2.3 | | 分子名称: | pRO-2.3 | | 著者 | Boyken, S.E, Sankaran, B, Bick, M.J, Baker, D. | | 登録日 | 2018-10-17 | | 公開日 | 2019-05-08 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.28 Å) | | 主引用文献 | De novo design of tunable, pH-driven conformational changes.
Science, 364, 2019
|
|
6N9H
 
 | | De novo designed homo-trimeric amantadine-binding protein | | 分子名称: | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine, SODIUM ION, amantadine-binding protein | | 著者 | Park, J, Baker, D. | | 登録日 | 2018-12-03 | | 公開日 | 2019-12-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.039 Å) | | 主引用文献 | De novo design of a homo-trimeric amantadine-binding protein.
Elife, 8, 2019
|
|
4A2S
 
 | | Structure of the engineered retro-aldolase RA95.5 | | 分子名称: | 1-(6-METHOXYNAPHTHALEN-2-YL)BUTANE-1,3-DIONE, INDOLE-3-GLYCEROL PHOSPHATE SYNTHASE | | 著者 | Giger, L, Caner, S, Kast, P, Baker, D, Ban, N, Hilvert, D. | | 登録日 | 2011-09-28 | | 公開日 | 2012-11-07 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Evolution of a designed retro-aldolase leads to complete active site remodeling.
Nat.Chem.Biol., 9, 2013
|
|
6E5C
 
 | | Solution NMR structure of a de novo designed double-stranded beta-helix | | 分子名称: | De novo beta protein | | 著者 | Marcos, E, Chidyausiku, T.M, McShan, A, Evangelidis, T, Nerli, S, Sgourakis, N, Tripsianes, K, Baker, D. | | 登録日 | 2018-07-19 | | 公開日 | 2018-11-07 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | De novo design of a non-local beta-sheet protein with high stability and accuracy.
Nat. Struct. Mol. Biol., 25, 2018
|
|
