7KHU
 
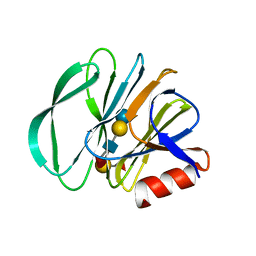 | | Crystal structure of the carbohydrate-binding domain VP8* of human P[4] rotavirus strain BM5265 in complex with LNDFH I | | Descriptor: | Outer capsid protein VP4, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Xu, S, Stuckert, M, Burnside, R, McGinnis, K, Jiang, X, Kennedy, M.A. | | Deposit date: | 2020-10-22 | | Release date: | 2021-08-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural basis of P[II] rotavirus evolution and host ranges under selection of histo-blood group antigens.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7KI5
 
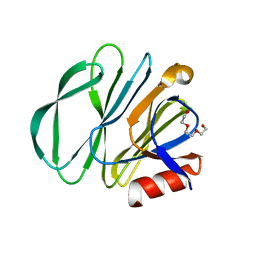 | | Crystal structure of P[6] rotavirus vp8* in complex with LNT | | Descriptor: | Capsid protein, GLYCEROL, TRIETHYLENE GLYCOL, ... | | Authors: | Xu, S, Kennedy, M.A. | | Deposit date: | 2020-10-23 | | Release date: | 2021-08-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural basis of P[II] rotavirus evolution and host ranges under selection of histo-blood group antigens.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6NIW
 
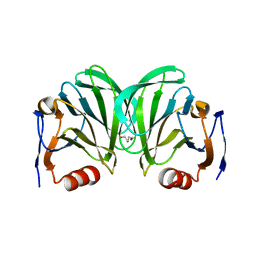 | | Crystal structure of P[6] rotavirus | | Descriptor: | DI(HYDROXYETHYL)ETHER, Protease-sensitive outer capsid protein | | Authors: | Xu, S, Liu, Y, Lakamp, L, Ahmed, L, Jiang, X, Kennedy, M.A. | | Deposit date: | 2019-01-01 | | Release date: | 2020-01-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Molecular basis of P[II] major human rotavirus VP8* domain recognition of histo-blood group antigens.
Plos Pathog., 16, 2020
|
|
1XHS
 
 | | Solution NMR Structure of Protein ytfP from Escherichia coli. Northeast Structural Genomics Consortium Target ER111. | | Descriptor: | Hypothetical UPF0131 protein ytfP | | Authors: | Aramini, J.M, Huang, Y.J, Swapna, G.V.T, Paranji, R.K, Xiao, R, Shastry, R, Acton, T.B, Cort, J.R, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-09-20 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of Escherichia coli ytfP expands the structural coverage of the UPF0131 protein domain family.
Proteins, 68, 2007
|
|
6OAI
 
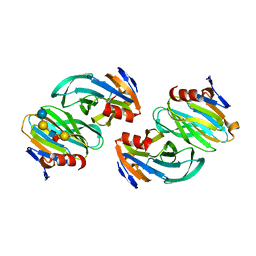 | | Crystal structure of P[6] rotavirus vp8* complexed with LNFPI | | Descriptor: | Protease-sensitive outer capsid protein, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Xu, S, Liu, Y, Jiang, X, Kennedy, M.A. | | Deposit date: | 2019-03-16 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis of P[II] major human rotavirus VP8* domain recognition of histo-blood group antigens.
Plos Pathog., 16, 2020
|
|
1XJS
 
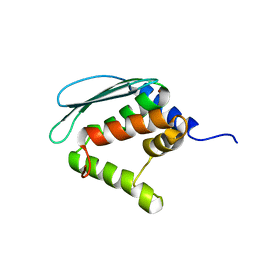 | | Solution structure of Iron-Sulfur cluster assembly protein IscU from Bacillus subtilis, with Zinc bound at the active site. Northeast Structural Genomics Consortium Target SR17 | | Descriptor: | NifU-like protein, ZINC ION | | Authors: | Kornhaber, G.J, Swapna, G.V.T, Ramelot, T.A, Cort, J.R, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-09-24 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Iron-Sulfur cluster assembly protein IscU
from Bacillus subtilis, with Zinc bound at the active site.
Northeast Structural Genomics Consortium Target SR17.
To be Published
|
|
1XPW
 
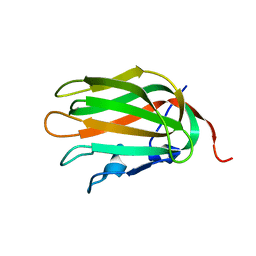 | | Solution NMR Structure of human protein HSPCO34. Northeast Structural Genomics Target HR1958 | | Descriptor: | LOC51668 protein | | Authors: | Ramelot, T.A, Xiao, R, Ma, L.C, Acton, T.B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-09 | | Release date: | 2004-11-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Improving NMR protein structure quality by Rosetta refinement: a molecular replacement study.
Proteins, 75, 2009
|
|
1XPN
 
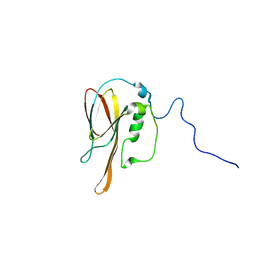 | | NMR structure of P. aeruginosa protein PA1324: Northeast Structural Genomics Consortium target PaP1 | | Descriptor: | hypothetical protein PA1324 | | Authors: | Cort, J.R, Ni, S, Lockert, E.E, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-08 | | Release date: | 2004-11-23 | | Last modified: | 2024-03-06 | | Method: | SOLUTION NMR | | Cite: | Structure and function of Pseudomonas aeruginosa protein PA1324 (21-170).
Protein Sci., 18, 2009
|
|
1YWW
 
 | |
1YWU
 
 | | Solution NMR structure of Pseudomonas Aeruginosa protein PA4608. Northeast Structural Genomics target PaT7 | | Descriptor: | hypothetical protein PA4608 | | Authors: | Ramelot, T.A, Yee, A.A, Cort, J.R, Semesi, A, Arrowsmith, C.H, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-02-18 | | Release date: | 2005-03-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure and binding studies confirm that PA4608 from Pseudomonas aeruginosa is a PilZ domain and a c-di-GMP binding protein.
Proteins, 66, 2007
|
|
1ZG2
 
 | | Solution NMR structure of the UPF0213 protein BH0048 from Bacillus halodurans. Northeast Structural Genomics target BhR2. | | Descriptor: | Hypothetical UPF0213 protein BH0048 | | Authors: | Aramini, J.M, Swapna, G.V.T, Xiao, R, Ma, L, Shastry, R, Ciano, M, Acton, T.B, Liu, J, Rost, B, Cort, J.R, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-04-20 | | Release date: | 2005-06-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the UPF0213 protein BH0048 from Bacillus halodurans. Northeast Structural Genomics target BhR2.
To be Published
|
|
1YX3
 
 | | NMR structure of Allochromatium vinosum DsrC: Northeast Structural Genomics Consortium target OP4 | | Descriptor: | hypothetical protein DsrC | | Authors: | Cort, J.R, Dahl, C, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-02-19 | | Release date: | 2005-04-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Allochromatium vinosum DsrC: solution-state NMR structure, redox properties, and interaction with DsrEFH, a protein essential for purple sulfur bacterial sulfur oxidation.
J.Mol.Biol., 382, 2008
|
|
1ZNO
 
 | | Crystal Structure of VC702 from Vibrio Cholerae, Northeast Structural Genomics Consortium Target: VcP1 | | Descriptor: | Hypothetical UPF0244 protein VC0702, MAGNESIUM ION | | Authors: | Ni, S, Forouhar, F, Bussiere, D.E, Robinson, H, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-05-11 | | Release date: | 2006-07-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of VC0702 at 2.0 A: conserved hypothetical protein from Vibrio cholerae.
Proteins, 63, 2006
|
|
1D1R
 
 | |
1EO1
 
 | |
1EIW
 
 | |
1EZE
 
 | | STRUCTURAL STUDIES OF A BABOON (PAPIO SP.) PLASMA PROTEIN INHIBITOR OF CHOLESTERYL ESTER TRANSFERASE. | | Descriptor: | CHOLESTERYL ESTER TRANSFERASE INHIBITOR PROTEIN | | Authors: | Buchko, G.W, Rozek, A, Kanda, P, Kennedy, M.A, Cushley, R.J. | | Deposit date: | 2000-05-10 | | Release date: | 2000-09-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural studies of a baboon (Papio sp.) plasma protein inhibitor of cholesteryl ester transferase.
Protein Sci., 9, 2000
|
|
1D4U
 
 | | INTERACTIONS OF HUMAN NUCLEOTIDE EXCISION REPAIR PROTEIN XPA WITH RPA70 AND DNA: CHEMICAL SHIFT MAPPING AND 15N NMR RELAXATION STUDIES | | Descriptor: | NUCLEOTIDE EXCISION REPAIR PROTEIN XPA (XPA-MBD), ZINC ION | | Authors: | Buchko, G.W, Daughdrill, G.W, de Lorimier, R, Rao, S, Isern, N.G, Lingbeck, J, Taylor, J, Wold, M.S, Gochin, M, Spicer, L.D, Lowry, D.F, Kennedy, M.A. | | Deposit date: | 1999-10-06 | | Release date: | 1999-10-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Interactions of human nucleotide excision repair protein XPA with DNA and RPA70 Delta C327: chemical shift mapping and 15N NMR relaxation studies.
Biochemistry, 38, 1999
|
|
1COC
 
 | | SOLUTION-STATE STRUCTURE OF A DNA DODECAMER DUPLEX CONTAINING A CIS-SYN THYMINE CYCLOBUTANE DIMER. | | Descriptor: | DNA (5'-D(*CP*TP*TP*AP*AP*TP*TP*CP*GP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*CP*GP*AP*AP*TP*TP*AP*AP*G)-3') | | Authors: | McAteer, K, Jing, Y, Kao, J, Taylor, J.-S, Kennedy, M.A. | | Deposit date: | 1999-05-26 | | Release date: | 1999-06-09 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution-state structure of a DNA dodecamer duplex containing a Cis-syn thymine cyclobutane dimer, the major UV photoproduct of DNA.
J.Mol.Biol., 282, 1998
|
|
1JI8
 
 | |
1L7Y
 
 | | Solution NMR Structure of C. elegans Protein ZK652.3. NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET WR41. | | Descriptor: | HYPOTHETICAL PROTEIN ZK652.3 | | Authors: | Cort, J.R, Chiang, Y, Zheng, D, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-03-18 | | Release date: | 2002-08-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of conserved eukaryotic protein ZK652.3 from C. elegans: a ubiquitin-like fold.
Proteins, 48, 2002
|
|
1JW3
 
 | | Solution Structure of Methanobacterium Thermoautotrophicum Protein 1598. Ontario Centre for Structural Proteomics target MTH1598_1_140; Northeast Structural Genomics Target TT6 | | Descriptor: | Conserved Hypothetical Protein MTH1598 | | Authors: | Chang, X, Connelly, G, Yee, A, Kennedy, M.A, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2001-09-02 | | Release date: | 2002-02-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An NMR approach to structural proteomics.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1RZW
 
 | | The Solution Structure of the Archaeglobus fulgidis protein AF2095. Northeast Structural Genomics Consortium target GR4 | | Descriptor: | Protein AF2095(GR4) | | Authors: | Powers, R, Acton, T.B, Huang, Y.J, Liu, J, Ma, L, Rost, B, Chiang, Y, Cort, J.R, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-12-29 | | Release date: | 2004-11-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Archaeglobus fulgidis peptidyl-tRNA hydrolase (Pth2) provides evidence for an extensive conserved family of Pth2 enzymes in archea, bacteria, and eukaryotes
Protein Sci., 14, 2005
|
|
1Q27
 
 | |
8E0N
 
 | |
